Department of Genetics, Rutgers University, Piscataway, NJ, USA
Departament de Biologia Cellular, Fisiologia i Immunologia, Universitat Autònoma de Barcelona (UAB), Barcelona, Spain
Department of Biological Sciences, Seton Hall University, South Orange, NJ, USA
Abstract
The mammalian genome encodes three Aurora protein kinase homologs (AURKA/B/C) which regulate chromosome segregation in nearly every cell type. AURKC expression is largely limited to meiotic cells. Because of the similarity in sequences between AURKB and AURKC, determining their separate functions during meiosis is challenging. We designed a chemical genetics approach to investigate AURKB function. Using Crispr/Cas9 genome editing in mouse, we replaced an ATP binding pocket amino acid to permit binding of cell-permeable ATP analogs. We also introduced a second site supressor mutation to tolerate the pocket enlargement. Heterozygous mice were fertile, but never produced homozygous analog-sensitive mice. Because Aurkb is an essential gene, we conclude that this analog-sensitive allele is either catalytically inactive or not fully catalytically active in mouse.
Description
The Aurora protein kinases (AURK) are essential regulators of chromosome segregation in mitotic and meiotic cell divisions (Carmena and Earnshaw 2003). Unlike mitotically dividing cells which express two AURKs (AURKA and AURKB), mammalian oocytes, which undergo meiosis, express three AURK homologs: AURKA/B/C (Brown et al. 2004; Nguyen et al. 2018). Because AURKC shares high sequence homology with AURKB, standard approaches such as RNAi knockdown (Chen et al. 2005; Sharif et al. 2010) and small molecule inhibition (Chen et al. 2005; Lane et al. 2010; Sharif et al. 2010; Shuda et al. 2009; Swain et al. 2008) to understand their roles do not provide the specificity required to assess non-overlapping functions. Furthermore, these kinases can compensate for one another in oocytes from mouse knockout strains (Balboula and Schindler 2014; Fernandez-Miranda et al. 2011; Nguyen et al. 2018; Schindler et al. 2012), making phenotype assessments less clear. Therefore, alternative strategies to provide specificity are needed.
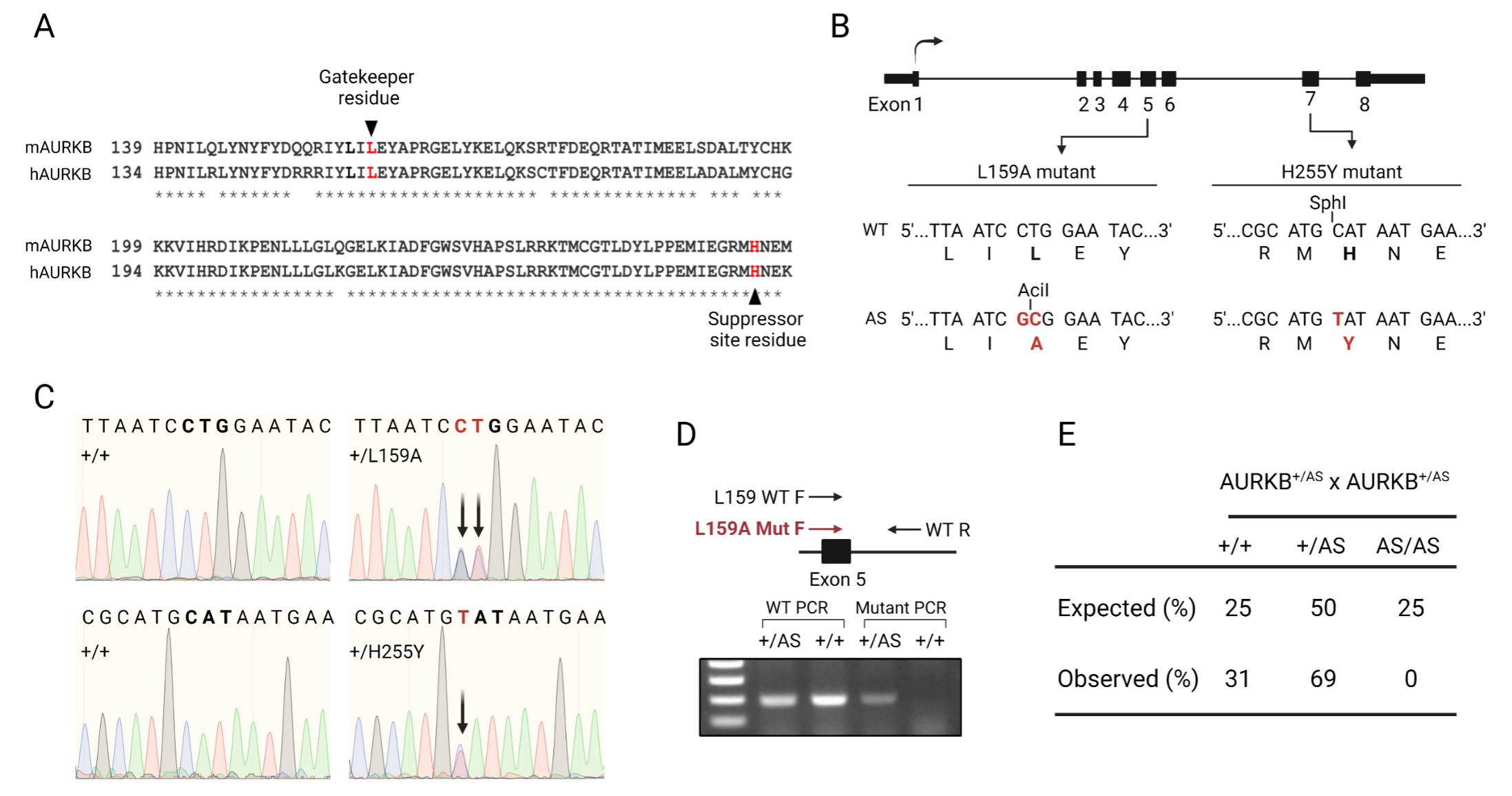
Chemical genetics is a strategy that documents superior specificity in inhibiting protein kinases. This approach involves enlarging the kinase ATP-binding pocket by introduction of an amino acid substitution. This enlargement allows specific acceptance of cell-permeable, bulky ATP analog inhibitors that cannot bind other kinases in the cell (Garske et al. 2011). Although most protein kinases tolerate the ATP pocket enlargement, approximately 30% of protein kinases do not (Zhang et al. 2005). A previous report demonstrates that human AURKB ATP-pocket enlargement (Leucine 154 to Alanine) abolishes its catalytic activity. Excitingly, this activity can be rescued by mutation of a second site suppressor in the C-terminus of the kinase (Histidine 250 to Tyrosine). When HeLa and U2OS cells were transiently transfected with the analog sensitive human AURKB allele, the kinase was active and cell division was not altered (Hengeveld et al. 2012). Based on these data, we were keen to adopt this approach to study the potential unique functions of AURKB. We used Crispr/Cas9 genome editing to introduce the ATP-binding pocket point mutation (L159 in mouse) and second site suppressor mutation (H255 in mouse) into the mouse genome (Fig. 1A). After confirmation by restriction digestion (Fig. 1B) and Sanger sequencing (Fig. 1C), we obtained 4 founders (2 male, 2 female) that were heterozygous for the analog-sensitive allele on the same chromosome. To expand the colony, we set up breeding cages using the heterozygous founders. Heterozygous animals were fertile and gave birth to pups. We also crossed heterozygous pups to increase chances of obtaining homozygous animals. After genotyping 80 pups that were born to 8 heterozygous mating pairs, we did not obtain any homozygous mice; wild-type and heterozygotes were born at altered Mendelian ratios (Fig. 1D-E). Because Aurkb is an essential gene (Fernández-Miranda et al. 2011), these data indicate that this analog-sensitive allele of Aurkb is either catalytically inactive or not fully catalytically active in mouse and that it cannot be used to determine specific AURKB functions in mouse oocytes.
Reagents
C57BL/6J mice: Jackson Laboratory
Cas9 IDT
L-A RNA guide (PAM sequence in parentheses) from Sigma: ACTACTTCTACGACCAGCAG (AGG)
L-A donor oligo (mutations in lower case):
CAGGCACTAGATCCCACACCAGGTAGGCCCTGACCGTGGCAGTCCGCTGCTCATCGAAGGTCCGACTCTTCTGCAGT
TCCTTGTAGAGTTCCCCGCGAGGGGCGTATTCCgcGATTAAGTAGATtCTCTGCTGGTCGTAGAAGTAGTTGTAGAG
TTGAAGGATGTTGGGATGTCTGG
H-Y RNA guide (PAM sequence in parentheses) from Sigma: TGCCCCCAGAGATGATTGAG (GGG)
H-Y donor oligo (mutations in lower case):
TCCGACGATACGTCTCACTGTGGCTAGGGCTCTCGAAGGGTGGGTTCCCCACCATCAGTTCATAGCAGAGCACCCC
GATGCACCATAGATCTACCATTTCATTATaCATGCGgCCCTCAATCATCTCTGGGGGCAGATAGTCCAGCGTGCCG
CACATGGTCTTCCTCCTGGTGGGATGAG
Genotyping Primers:
L159A substitution
L159WT F: 5’- CCAGCAGAGGATCTACTTAATCCT -3’
L159AS F: 5’- CCAGCAGAGGATCTACTTAATCGC -3’
WT R: 5’- ATGATctgtgagggtccacacaag -3’
H255Y substitution
H255WT F: 5’- GAGATGATTGAGGGGCGCATGCAT -3’
H255Y F: 5’- GAGATGATTGAGGGGCGCATGTAT -3’
WT R: 5’- CTGGCTCCTACAGTACACAAAGG -3’
Acknowledgments
The authors acknowledge the efforts of Drs. Peter Romenienko and Ghassan Yehia at the Rutgers Cancer Institute of New Jersey Genome Editing Shared Resource for designing, testing and creating these modified mice. We also acknowledge efforts of Ms. Christine Prorock-Rogers in mouse colony husbandry and genotyping.
References
Funding
This project was supported by funding from the National Institutes of Health: R01 GM112801 to KS. Services, results and/or products in support of the research project were generated by the Rutgers Cancer Institute of New Jersey Genome Editing Shared Resource, supported, in part, with funding from NCI-CCSG P30CA072720-5922.
Reviewed By
AnonymousHistory
Received:Accepted: October 18, 2021
Published: November 23, 2021
Copyright
© 2021 by the authors. This is an open-access article distributed under the terms of the Creative Commons Attribution 4.0 International (CC BY 4.0) License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.Citation
Vazquez, BN; Quartuccio, SM; Schindler, K (2021). An analog-sensitive allele of Aurora kinase B is lethal in mouse. microPublication Biology. 10.17912/micropub.biology.000491.Download: RIS BibTeX