Abstract
In healthy eukaryotic cells, the two leaflets that make up plasma membranes are highly asymmetric with respect to the lipids they contain. In both unicellular eukaryotes and metazoans, the asymmetry in the distribution of aminophospholipids is maintained by P4-family transmembrane ATPases, which catalyze the movement of selected phospholipids from the outer leaflet to the inner. C. elegans has six P4-family ATPases, TAT-1 – TAT-6. TAT-1 – TAT-5 are expressed in many tissues and cells. Here we report that, in contrast, TAT-6 is much less broadly expressed and that, within the somatic gonad, expression of TAT-6 reporters is restricted to the spermathecal-uterine core cell (sujc) cells.
Description
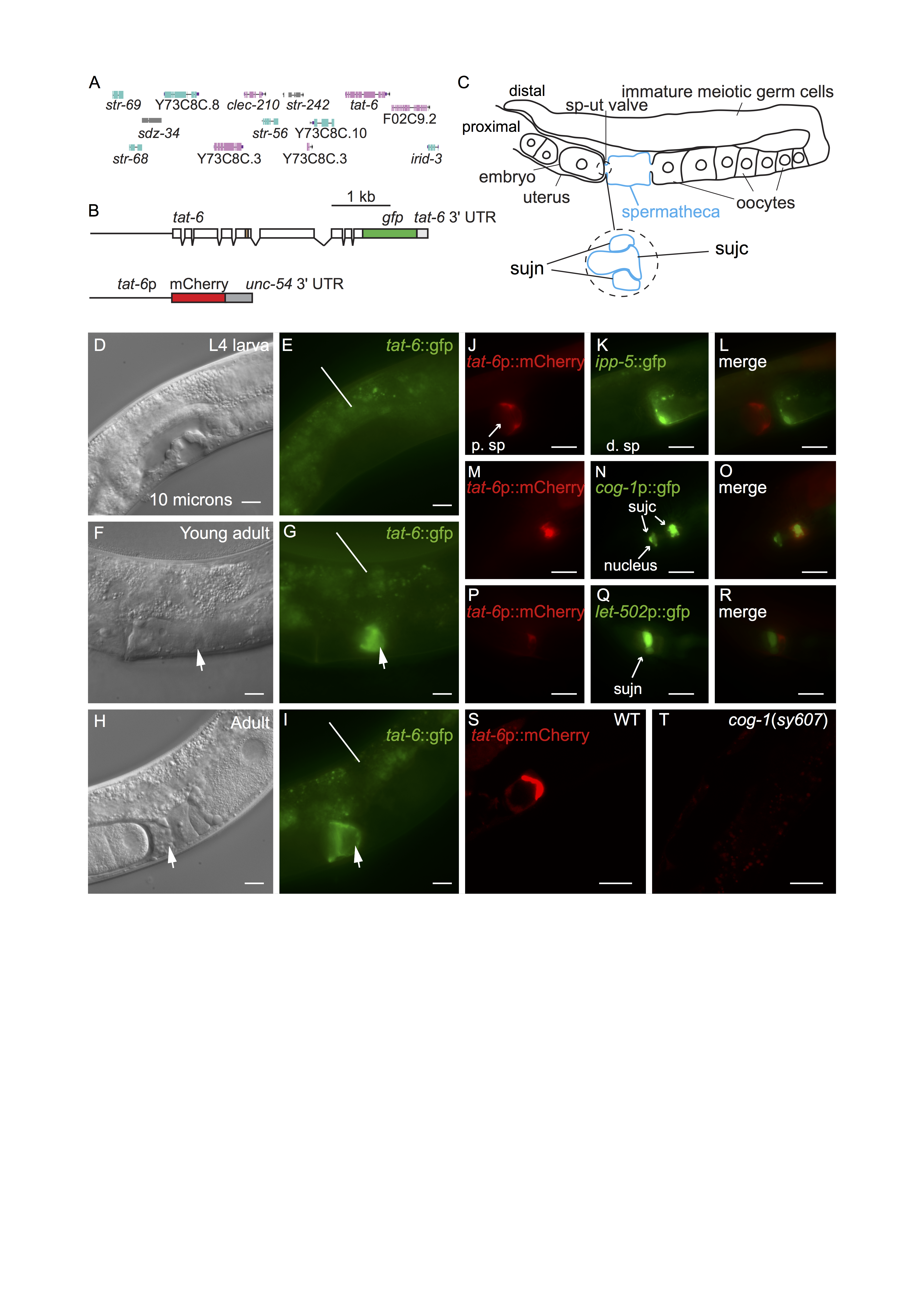
tat-1 and tat-5 are both very broadly expressed in C. elegans (Lyssenko et al. 2008, Ruaud et al. 2009, Chen et al. 2010, Wehman et al. 2011). tat-2, tat-3 and tat-4 are also expressed in many cells and tissues although apparently not ubiquitously (Lyssenko et al. 2008). No reports exist presently describing the pattern in which tat-6 is expressed. We generated multiple transgenic lines containing a construct encoding the entire TAT-6 protein fused to GFP (Fig. 1A,B). The construct contained tat-6 promoter sequences, all exons and introns as well as intragenic sequences to the left and right of the tat-6 coding region (Fig. 1A,B) (Sarov et al. 2012). Expression was seen in cells in the head and tail but in the center of the worm, expression was restricted to a proximal part of the gonad (Fig. 1C,F,G,H,I). Expression in the gonad was absent in early and mid-L4 stage worms but was robust in young adults (Fig. 1D,E,F,G). Prior to ovulation, fluorescence was seen in a region at the junction between the spermatheca and the uterus (Fig. 1C,F,G). In hermaphrodites in which ovulation had occurred, GFP fluorescence was seen partially surrounding the distal-most egg in the uterus (Fig.1H,I). The valve forming the junction between spermatheca and the uterus consists of a toroidal syncytium, sujn, and a core cell syncytium, sujc, which initially occupies the center of the valve (Fig. 1C) (Kimble and Hirsh 1979, Lints and Hall 2013). During the first ovulation, the core is displaced by the passage of the newly fertilized egg from the spermatheca to the uterus (Kimble and Hirsh 1979). The change in the distribution of TAT-6::GFP fluorescence we observed during the first ovulation suggested that TAT-6 might be expressed in sujc. Since existing GFP markers for cells in the spermatheca were available, to determine in which cells tat-6 was expressed, we first constructed strains containing a tat-6p::mCherry transcriptional reporter (Fig. 1B). The marker was expressed in the same way as the GFP reporter (Fig. 1S). In a strain containing the tat-6p::mCherry reporter and a reporter in which GFP expression was driven by promoter sequences from the cog-1 gene active in sujc (Palmer et al. 2002), the mCherry and GFP signals were seen in the same cell (Fig. 1M,N,O). In contrast, in a strain containing the tat-6p::mCherry reporter and an ipp-5p::gfp reporter, which is expressed in distal spermathecal cells that form part of the junction with the ovary (Bui and Sternberg 2002), the two fluorescent signals did not overlap (Fig. 1J,K,L). Similarly, in a strain containing the tat-6p::mCherry reporter and an let-502p::gfp reporter, which is strongly expressed in sujn cells (Wissmann et al. 1999), the mCherry signal was adjacent to the strong GFP expression rather than coincident with it (Fig. 1P,Q,R). To further verify the identity of the cells expressing the tat-6 reporters, we crossed the tat-6p::mCherry transgene into a cog-1(sy607) mutant background. cog-1 encodes a GTX/Nkx6.1 homeodomain transcription factor; in cog-1(sy607) mutant hermaphrodites, cells having the morphology of sujc cells are absent (Palmer et al. 2002). cog-1 is not expressed in sujn cells (Palmer et al. 2002). Consistent with the results with the fluorescent markers, no tat-6 reporter expression was seen in the gonad in the cog-1(sy607) mutant (Fig. 1S,T).
We do not presently know the function of TAT-6 in sujc. Indeed, the function of the sujc cells themselves is not presently known: ablation of sujc cells in the mid-L4 stage causes only a weak effect on brood size (Palmer et al. 2002) (although an earlier function for sujc cells in morphogenesis of the spermathecal-uterine junction has not been ruled out (Palmer et al. 2002)). Genetic research in Saccharomyces cerevisiae has revealed that all five P4-family ATPases in this organism, through their actions as phospholipid translocases, promote one or more vesicle transport events in the endosomal or secretory pathways (Hankins et al. 2015, Pomorski and Menon 2016, Yang et al. 2018). It is thought that aminophospholipid translocases promote membrane bending that occurs during the formation of transport vesicles (Hankins et al. 2015, Pomorski and Menon 2016, Yang et al. 2018). C. elegans tat-1 is required for correct vesicle transport within the endolysosomal system (Ruaud et al. 2009, Chen et al. 2010, Nilsson et al. 2011). C. elegans tat-5 is required for endosome to Golgi trafficking of MIG-14 (a C. elegans Wntless homologue) in the QL neuroblast (McGough et al. 2018), and to suppress the formation of extracellular vesicles in the embryo (Wehman et al. 2011, Beer et al. 2018). Thus, it is possible that TAT-6 promotes one or more vesicle transport event within sujc. It is worth noting that the plasma membrane of sujc cells is unusual in being highly convoluted (Kimble and Hirsh 1979, Lints and Hall 2013). Although it is not known how the extensive folding of the membrane arises, cell autonomous processes that promote membrane folding in sujc cells have not been ruled out. Finally, our studies also shed light on what happens to material from the sujc cells following ovulation. The fate of these cells after the core of the spermathecal-uterine valve has been displaced is presently not known. The fact that some TAT-6::GFP fusion protein (and mCherry) remains in the uterine epithelium even in older hermaphrodites indicates that at least a part of the sujc cells is retained following ovulation.
Methods
Request a detailed protocolStandard methods were used in the maintenance of C. elegans worms. Clone I16253250892533I A08 (Sarov et al. 2012) was used to generate svEx940, the extrachromosomal array from which the integrated array svIs144 was derived; pVB652 was used to generate svEx967 and svEx968. Transgenic strains were generated by microinjection (Fire 1986). The DNA clones were microinjected at a concentration of 50 ng/μl. svIs144 was derived from svEx940 by γ-irradiation. Micrographs were made with DM6000 B and DMRB compound microscopes (Leica); confocal micrographs were made with an A1 confocal microscope (Nikon).
Reagents
| Strain | Genotype | Available from |
| VB3030 | unc-119(ed3op); svIs144[tat-6p::tat-6::GFP unc-119(+)] | This work |
| VB3041 | unc-4(e120); svEx967[tat-6p::mCherry unc-4(+)] | This work |
| PS3747 | ipp-5(sy605); syEx429[ipp-5::GFP rol-6(su1006)] | CGC |
| PS3662 | syIs63[cog-1p::GFP] | CGC |
| VB2238 | svEx968[tat-6p::mCherry]; syEx429[ipp-5::GFP rol-6(su1006)] | This work |
| VB3122 | syIs63[cog-1p::GFP]; svEx968[tat-6p::mCherry unc-4(+)] | This work |
| HR606 | sbEx136[let-502p::GFP rol-6(su1006)] | CGC |
| VB3123 | sbEx136[let-502p::GFP rol-6(su1006)]; svEx968[tat-6p::mCherry unc-4(+)] | This work |
| VB3357 | cog-1(sy607); svEx967[tat-6p::mCherry unc-4(+)] | This work |
References
Funding
Cancerfonden grant 14683 to ST
Reviewed By
AnonymousHistory
Received: September 6, 2021Revision received: October 25, 2021
Accepted: October 25, 2021
Published: November 4, 2021
Copyright
© 2021 by the authors. This is an open-access article distributed under the terms of the Creative Commons Attribution 4.0 International (CC BY 4.0) License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.Citation
Nilsson, L; Rahmani, S; Tuck, S (2021). C. elegans TAT-6, a putative aminophospholipid translocase, is expressed in sujc cells in the hermaphrodite gonad.. microPublication Biology. 10.17912/micropub.biology.000495.Download: RIS BibTeX