European Molecular Biology Laboratory, European Bioinformatics Institute, Wellcome Trust Genome Campus, Hinxton, Cambridge CB10 1SD, UK
Abstract
C. elegans was the first animal to have its genome completely sequenced. In the decades since, the genome continues to be actively curated, annotated, and improved. Here we report the discovery of a new gene in a region of the genome that is currently not associated with any annotated gene or feature. We present RNA-seq and RT-PCR evidence that this gene is expressed at detectable levels, and that it is alternatively spliced. The new gene (Y97E10C.2) shares an operon with two upstream genes. We provide RNA-seq and RT-PCR evidence for a missing exon in the upstream gene T05B11.7, as well as an alternatively-spliced exon.
Description
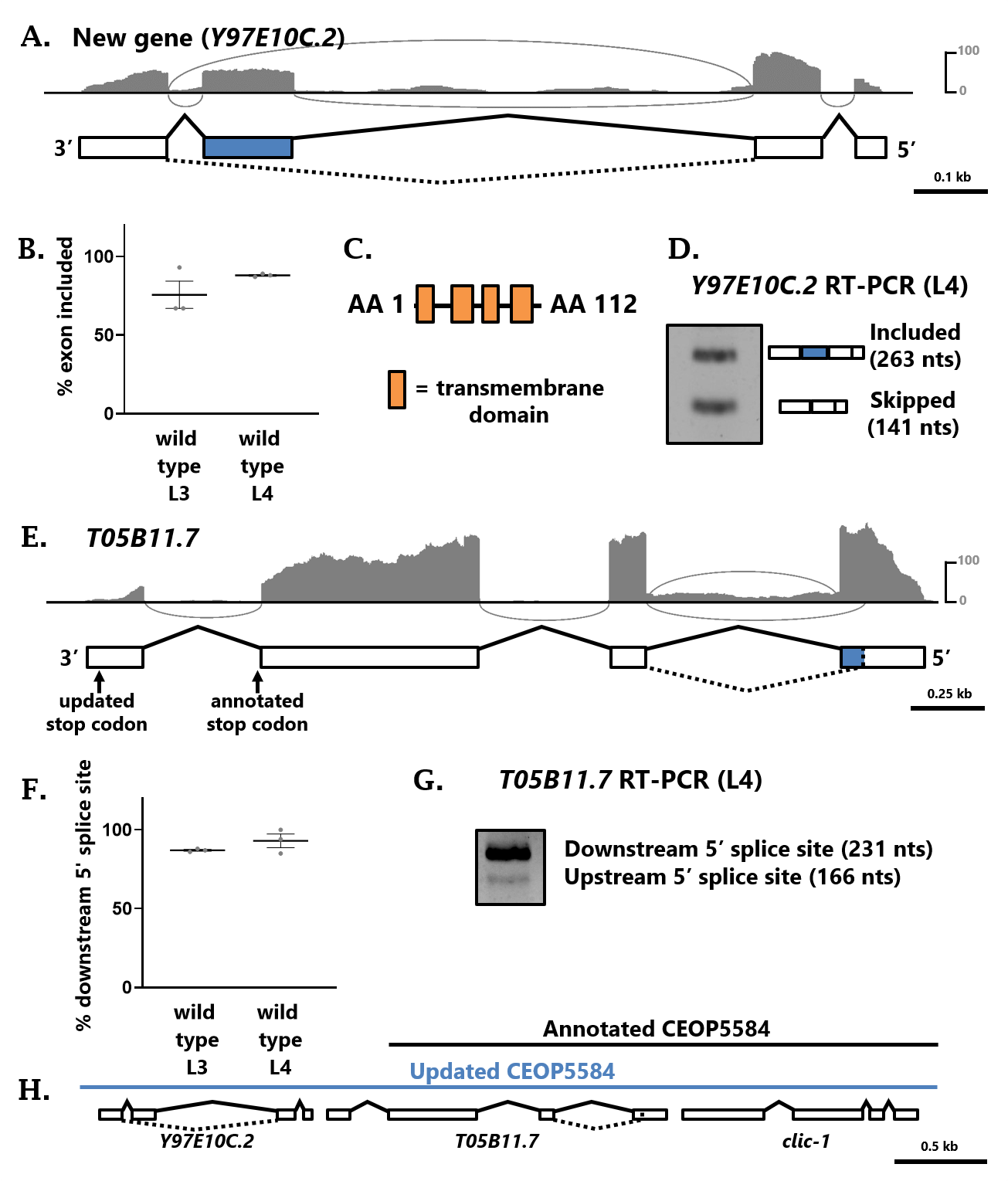
In the decades since the C. elegans genome sequence was completed (C. elegans Sequencing Consortium, 1998), ongoing curation efforts continue to further improve and annotate the genome (Harris et al., 2020). In the course of searching for alternatively-spliced genes using RNA-seq data, we happened upon an alternative splicing event in a region of the C. elegans genome with no associated gene or feature annotation. Upon closer inspection, we identified RNA-seq support for an unannotated four-exon gene, with the third exon being alternatively spliced (Fig 1A). The putative coding sequences of this gene encode proteins of either 86 or 122 amino acids, depending on the splicing choice. The longer isoform (exon three included) is the major isoform according to our RNA-seq data (Fig 1B), and this isoform encodes four predicted transmembrane domains (Fig 1C). RT-PCR of wild-type L4 stage worms further confirms that the gene is expressed at detectable levels, and that exon three is alternatively spliced (Fig 1D). We have submitted this information to WormBase, and the new gene is set to appear as WBGene00306123 (transcripts Y97E10C.2a and Y97E10C.2b) in release WS283.
While inspecting this locus, we also noted RNA-seq evidence for the gene immediately upstream (T05B11.7) that deviates from its annotated gene model. The RNA-seq data indicate that prior to reaching the annotated stop codon in exon three, the mRNA is spliced to an unannotated downstream exon (Fig 1E). Therefore, the annotated stop codon is in fact located within an intron, and the T05B11.7 gene contains four exons. This increases the coding sequence of T05B11.7 from 852 to 924 nucleotides. We also found evidence for alternative splicing in the T05B11.7 gene. Exon one harbors an alternative 5’ splice site which is used in a small fraction of the transcripts detected by RNA-seq (Fig 1F) and which is also detectable via RT-PCR (Fig 1G). These alternative isoforms (including the new addition of exon four) will appear in WS283 as transcripts T05B11.7a and T05B11.7b.
Finally, the gene T05B11.7 is currently annotated as part of an operon (CEOP5584) along with its upstream gene clic-1 (Fig 1H). Our newly-described gene Y97E10C.2, which is located immediately downstream of T05B11.7, is now included as the final gene in this operon, making CEOP5584 a 3-gene operon (Fig 1H).
Methods
Request a detailed protocolAnalysis of RNA-seq data: Wild-type RNA-seq data used in this manuscript were previously published (Choudhary et al., 2021; Norris et al., 2017, 2014) using polyA-selected RNA obtained from either L3-stage or L4-stage wild-type (N2) hermaphrodites. In our initial search for alternative splicing which led to the identification of gene Y97E10C.2, we performed differential splicing analysis using JUM (Wang and Rio, 2018) on STAR-mapped (Dobin et al., 2013) short read (150 bp paired end) sequencing, stipulating that to be considered an alternative splicing event, both alternative junctions must be represented by at least 5 unique junction-spanning reads in at least two out of the three biological wild-type replicates tested.
RT-PCR: RT-PCR was performed on total RNA extracted from L4-stage wild-type (N2) hermaphrodites. Primers for Y97E10C.2: CGAGTCAAACTGAGCATGTG & AACACTCCACCAACAAGTAGAC. Primers for T05B11.7: TGTGCAACGGCAGCAAGAAG & CCAACAGTTTCCAGCTCCGAATC.
References
Funding
Research reported in this publication was supported by the National Institute of General Medical Sciences of the National Institutes of Health under award number R35GM133461, as well as the Welch Foundation (N-2042-20200401).
Reviewed By
Teresa Lee and Paul DavisHistory
Received: October 19, 2021Revision received: October 26, 2021
Accepted: October 27, 2021
Published: November 3, 2021
Copyright
© 2021 by the authors. This is an open-access article distributed under the terms of the Creative Commons Attribution 4.0 International (CC BY 4.0) License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.Citation
Iyengar, A; Diamantakis, S; Norris, A (2021). A new gene on C. elegans chromosome V. microPublication Biology. 10.17912/micropub.biology.000496.Download: RIS BibTeX