Present affiliation: Oncology Genomics Department, Diagnostica Longwood S.L. 50011 Zaragoza, Spain
Present affiliation: Department of Medical and Molecular Genetics, King’s College London, London SE1 9RT, UK
Present affiliation: Boston University School of Medicine, Department of Biochemistry, Genome Science Institute, Boston, MA 02118, USA
Abstract
The only representative of the MYC superfamily transcription factors in C. elegans, MML-1 (Myc and Mondo-like 1), was shown to promote extended lifespan in a variety of models and to regulate some aspects of C. elegans development. This previous research did not involve molecular characterization of MML-1. Here we use available mml-1 mutant alleles and other reagents to demonstrate that MML-1 is modified by O-GlcNAc, binds to promoters of some genes directly regulated by the DOT-1.1 histone methyltransferase complex, and has a role in promoting neuronal migration. Surprisingly, we found that the deletion allele mml-1(ok849), which was considered a null, produces an internally truncated protein resulting from an in-frame deletion. Localization of this truncated product to MML-1 target promoters was not impaired. The deleted region of MML-1 is proline-rich, and its function is poorly understood in mammalian homologs of MML-1. Based on our work and previously published data we conclude that the internal proline-rich region of MML-1 is dispensable for DNA binding but is biologically important.
Description
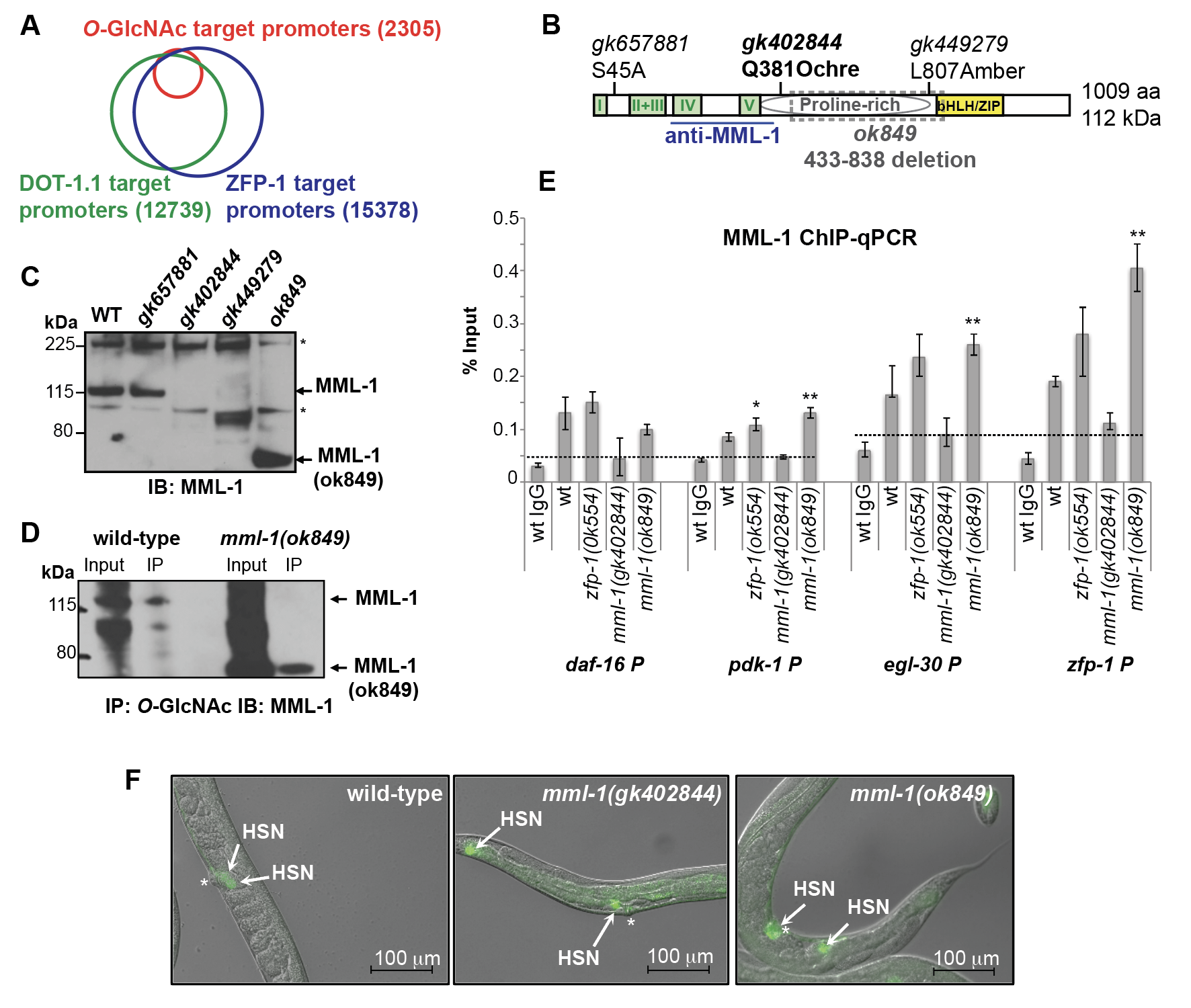
The predominant and ubiquitously expressed C. elegans histone H3 lysine 79 methyltransferase DOT-1.1 and its interacting partner, ZFP-1, play important roles in negatively modulating widely and highly expressed genes through promoter binding (Cecere et al. 2013) and in creating permissive environment for expression of developmentally regulated genes through enhancers (Esse et al. 2019; Esse and Grishok 2020). However, the mechanistic aspects of the ZFP-1/DOT-1.1 complex function are not clear. This study was motivated by the observation that genome-wide promoter-associated ZFP-1 and DOT-1.1 chromatin localization peaks (Mansisidor et al. 2011; Cecere et al. 2013) overlap with those of the O-GlcNAc modification on chromatin observed in wild type C. elegans (Love et al. 2010) (Figure 1A). Notably, the identities of the O-GlcNAc-modified proteins contributing to the O-GlcNAc ChIP-chip signals at the select promoters remain unknown. We chose to investigate the C. elegans basic helix- loop-helix leucine-zipper (bHLH/ZIP) transcription factor MML-1 (Myc and Mondo-like 1) (Pickett et al. 2007; Grove et al. 2009) as a candidate O-GlcNAc-modified protein, because both c-Myc (Chou et al. 1995) and the Mondo B protein (i.e., the Carbohydrate Response Element Binding Protein (ChREBP)) (Guinez et al. 2011) carry O-GlcNAc modification. MML-1 is a 1009 amino acid protein with multiple conserved domains, including the N-terminal Mondo Conserved Regions (I-V) (Pickett et al. 2007), which form a glucose sensing module in ChREBP (Abdul-Wahed et al. 2017), a proline-rich region, similar to that in ChREBP (Ortega-Prieto and Postic 2019), and a C-terminal DNA-binding and MXL-2 heterodimerization region called bHLH/ZIP (Pickett et al. 2007; Grove et al. 2009) (Figure 1B).
We obtained several viable mml-1 mutant alleles generated by the Million Mutation Project (Figure 1B, C) (Thompson et al. 2013). These included the potential null mutant mml-1(gk402844), which lacks the MML-1 protein signal recognized by the N-terminus-specific anti-MML-1 antibody (Figure 1C) (Pickett et al. 2007) (a generous gift from Dr. Donald Ayer). Notably, the mml-1(ok849) deletion allele (Figure 1B), which had been considered null (Johnson et al. 2014; Botts et al. 2016; Nakamura et al. 2016), leads to an in-frame deletion and produces an internally truncated protein (Figure 1C, D). We found that the O-GlcNAc-specific antibody RL2 immunoprecipitated both the wild type MML-1 and MML-1(ok849) (Figure 1D), which is consistent with our hypothesis that MML-1 is O-GlcNAc-modified. Moreover, both the wild type MML-1 and the MML-1(ok849) binding was detected at the promoters of the previously annotated ChIP targets of ZFP-1 (Mansisidor et al. 2011; Cecere et al. 2013) (Figure 1E). Notably, mml-1(gk402844) showed a background ChIP signal, whereas an increase in ChIP signal compared to that in wild type worms has been observed in mml-1(ok849) (Figure 1E). We believe that this increased promoter occupancy by MML-1(ok849) is due to the increased protein stability (Figure 1C, D), which suggests a role of the deleted region in regulating MML-1 dynamics.
Our interest in MML-1 has been driven by the hypothesis that MML-1 and ZFP-1/DOT-1.1 co-regulate common target genes. Since mml-1(ok849) phenocopied mml-1(RNAi) and behaved like a strong loss-of-function in published functional studies (Johnson et al. 2014; Nakamura et al. 2016), we were surprised to detect the truncated product by western immunoblotting (Figure 1C). However, the current annotation of the mml-1(ok849) allele is correct and predicts an in-frame deletion in MML-1, which results in a 608 aa long MML-1(ok849) (~67 kDa). This predicted size is consistent with our western data (Figure 1C, D).
To gain additional evidence that mml-1(ok849) and mml-1(gk402844) (a null) shared loss-of-function phenotypes, we analyzed Hermaphrodite-Specific Neuron (HSN) migration. The HSN under-migration phenotype is observed in zfp-1(ok554) reduction-of-function mutant, and it is, at least in part, due to the mis-regulation of pdk-1 expression in the hypodermis (Kennedy and Grishok 2014). Indeed, both mml-1(ok849) and mml-1(gk402844) worms displayed defects in HSN migration, 8% (n=49) and 45% (n=49), respectively (Figure 1F), analogous to those that we previously described for zfp-1(ok554) (25-30% affected animals) (Kennedy and Grishok 2014). We note that the mml-1(ok849) mutant animals exhibited both a less severe and a less penetrant HSN migration phenotype compared to the mml-1(gk402844) null. Interestingly, the MML-1/MXL-2 complex has earlier been implicated in cell migration in the male tail, and it acted non-autonomously, in the hypodermis, to promote cell migration (Pickett et al. 2007). In sum, these results suggest that MML-1/MXL-2 and ZFP-1/DOT-1.1 may act together in regulating genes expressed in the hypodermis to non-autonomously promote cell migrations during C. elegans development.
The MML-1 region deleted in MML-1(ok849) is proline-rich and similar to the proline-rich region in ChREBP, whose function is poorly understood (Ortega-Prieto and Postic 2019). Our findings, together with the previous studies of lifespan control using mml-1(ok849) and mxl-2(tm1516), which showed similar phenotypes and behaved like strong loss-of-function mutations (Johnson et al. 2014; Nakamura et al. 2016), indicate the importance of the proline-rich domain in the biological functions of MML-1. Thus, the existing data indicate that the MML-1(ok849) protein is less active, despite its binding to DNA and its O-GlcNAc modification. Moreover, they suggest that a posttranslational activation step targeting the proline-rich region of MML-1, and possibly ChREBP, is required for these DNA-binding proteins to be fully functional in transcription regulation.
Methods
Request a detailed protocolWestern immunoblotting
Approximately 20-25 L3 stage worms were picked directly into the loading buffer and boiled before loading. Proteins were resolved on precast NuPAGE Novex 4–12% Bis-Tris gels (Invitrogen) and transferred to a nitrocellulose membrane (0.45 μm) by semidry transfer (BioRad Trans-Blot SD transfer cell) at a constant current of 0.12A for 1 hour. The blotted membranes were blocked with the blocking buffer (5% non-fat dry milk in TBS-T) at room temperature for 1 hour. Subsequently, they were incubated with the primary antibody overnight at 4°C and with the secondary antibody for 2 hours at room temperature. Three washes with TBS-T buffer were made between and after the incubation with the antibodies. The anti-MML-1 rabbit antiserum raised against the 189-376 aa region was a gift from Dr. Don Ayer, University of Utah. It was used at 1:5000 dilutions.
O-GlcNAc immunoprecipitation/ MML-1 western
Synchronized populations of C. elegans were grown on agarose 15 cm Petri dishes until larval stage three at 20°C with about 100,000 worms plated per dish. Worms were washed off the plates using isotonic M9 solution and pelleted. The pellet was resuspended in ice-cold RIPA buffer (10 mM Tris-HCl pH 8.0, 0.1% SDS, 1% Triton X-100, 0.1% sodium deoxycholate, 1 mM EDTA, 0.15 M NaCl) supplemented with Halt™ Protease Inhibitor Cocktail (Thermo Fisher Scientific) and sonicated with 30 sec pulses 10 times using Branson microtip sonicator at 10% power in the cold room. The RIPA buffer was used to limit co-immunoprecipitation of proteins not modified by O-GlcNAc. Protein extracts were span at 12,000 rpm at 4°C for 10 minutes. Protein concentration was quantified by the Bradford assay, and 8 μg of the anti-O-GlcNAc monoclonal antibody (clone RL2, Abcam ab2739) was used with 2.5 mg protein extract in each immunoprecipitation reaction. After 1 hour incubation (4°C) of the immunocomplex, 50μl of Dynabeads™ Protein G suspension (Thermo Fisher Scientific) was added, and the mixture was incubated at 4°C for 1-3 hours on rotation. Beads were recovered with a Dynal® Magnetic Particle Concentrator (Invitrogen). After four washes with the extraction buffer the beads were resuspended in 50μl of gel loading buffer and incubated at 95°C for 5 minutes. Western immunoblotting with the anti-MML-1 rabbit antiserum was performed as described above.
MML-1 ChIP/qPCR
Chromatin immunoprecipitation was performed as described in our earlier publications (Mansisidor et al. 2011; Cecere et al. 2012; Cecere et al. 2013; Gushchanskaia et al. 2019). Briefly, 2.5–3mg of the 2% paraformaldehyde cross-linked protein extract from L3 worms was incubated for 1h at 4°C with the specific antibody and the immune complexes were then incubated with 60 μl Dynabeads™ Protein G suspension (Thermo Fisher Scientific) for 1h at 4°C. 1 μl of the anti-MML-1 antiserum was used in each immunoprecipitation reaction. DNA was cleaned up with the Qiagen PCR purification kit. The immunoprecipitated DNA was quantified by qPCR using the ΔΔCt method to calculate the percentage of immunoprecipitation relative to the input.
ChIP/chip data overlap analysis
Genomic regions representing ZFP-1 and DOT-1.1 enrichment were fetched from the modENCODE server (http://www.modencode.org). O-GlcNac peaks were obtained from NCBI GEO (GSE18611). For identification of target genes, C. elegans transcription unit coordinates were extracted from the UCSC genome browser (RefSeq based on WS220/ce10). A transcription unit was called bound by ZFP-1, DOT-1.1 or O-GlcNac if its 1500 bp upstream regions overlapped with a ChIP peak. Overlap analysis was performed in R using the package valr (Riemondy et al. 2017).
Visualization of C. elegans HSN neurons
HSN neurons were detected by using the zdIs13[tph-1::gfp] transcriptional reporter, as previously described (Kennedy and Grishok 2014).
Reagents
C. elegans strains
| Strain Name | Genotype | Source |
| N2 | Wild type Bristol N2 | CGC |
| RB954 | mml-1(ok849) III | CGC |
| RB774 | zfp-1(ok554) III | CGC |
| AGK760 | mml-1(gk402844) III. 4x outcrossed from VC20787 | AGK lab |
| SK4013 | zdIs13[tph-1p::gfp] IV | CGC |
| AGK777 | mml-1(ok849) III; zdIs13[tph-1p::gfp] IV | AGK lab |
| AGK778 | mml-1(gk402844) III; zdIs13[tph-1p::gfp] IV | AGK lab |
| VC20787 | Million Mutations Project strain, contains mml-1(gk402844) III | CGC |
| VC40479 | Million Mutations Project strain, contains mml-1(gk657881) III | CGC |
| VC30257 | Million Mutations Project strain, contains mml-1(gk449279) III | CGC |
Antibodies
| Name | Source | Reference |
| anti-O-GlcNAc, RL2, mouse monoclonal | Abcam ab2739 | (Love et al. 2010) |
| anti-MML-1 antiserum, rabbit, raised against 189-376 aa | Ayer lab (U of Utah) | (Pickett et al. 2007) |
Primers used in ChIP/qPCR
| Name | Sequence |
| pdk-1P (F) | AAACAACACATAGACTTGTGCC |
| pdk-1P (R) | GTACGGTTGTTATCGCTTTCAG |
| egl-30P3 (F) | CGTGAGTTTGAGTGTCTCTG |
| egl-30P3 (R) | GATTAGGTTGTGGCTTTCCC |
| zfp-1P (F) | GTCGTCAATTCTATTTCTCGT |
| zfp-1P (R) | GATAGTAGCCGAAAGGAACAG |
| daf-16P (F) | GATTCTCCCTCTCCGTTCAC |
| daf-16P (R) | GTGATGAAGAAGGTGGTCTC |
Acknowledgments
The anti-MML-1 antiserum was a gift from Dr. Donald Ayer (University of Utah).
References
Funding
This research was financially supported by the National Institutes of Health (NIH) award (P30DK063608) to Columbia University, the Schaefer Research Scholars Program Award to AG, and Boston University startup funds (AG). AC was partially supported by an NIH Training grant – “Hormones: Biochemistry and Molecular Biology” (T32DK07328). Strains were provided by the C. elegans Gene Knockout Project at the Oklahoma Medical Research Foundation and by the C. elegans Reverse Genetics Core Facility at the University of British Columbia, which are part of the International C. elegans Gene Knockout Consortium. Some strains used in this study were obtained from the Caenorhabditis Genetics Center, which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440).
Reviewed By
AnonymousHistory
Received: August 30, 2021Revision received: October 31, 2021
Accepted: November 2, 2021
Published: November 9, 2021
Copyright
© 2021 by the authors. This is an open-access article distributed under the terms of the Creative Commons Attribution 4.0 International (CC BY 4.0) License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.Citation
Ceballos, A; Esse, R; Grishok, A (2021). The proline-rich domain of MML-1 is biologically important but not required for localization to target promoters. microPublication Biology. 10.17912/micropub.biology.000498.Download: RIS BibTeX