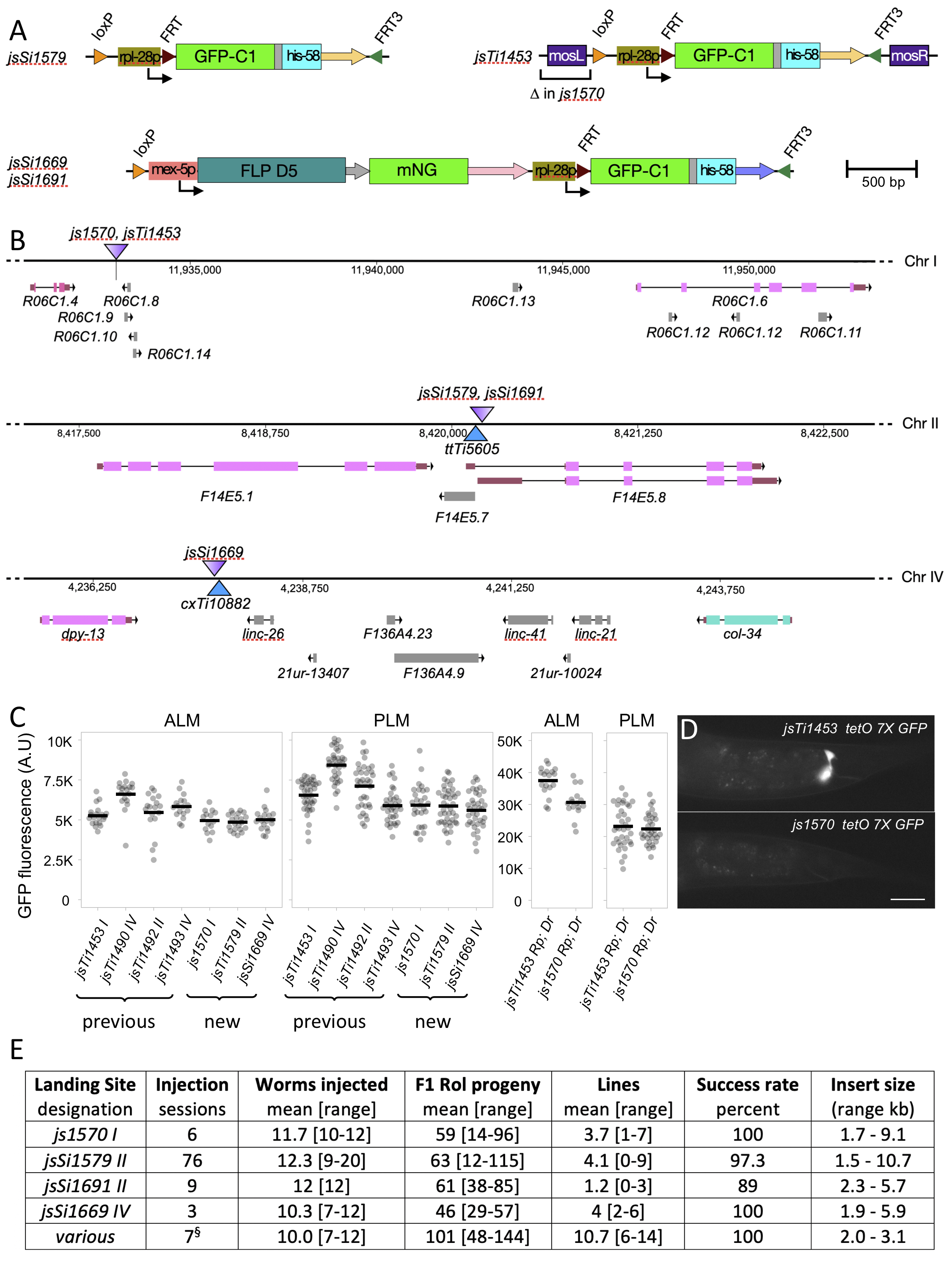
Abstract
Recombination-mediated cassette exchange (RMCE) is a recently developed alternative method for creating single copy transgenes using recombination rather than repair of double stranded breaks as the mechanism for driving integration into the genome. Two alternative methods for performing RMCE have been developed; a two-component approach using an unlinked source of FLP recombinase, and a one-component approach using a FLP expression cassette within the landing site. Here, I describe new landing sites for performing both types of RMCE. The new landing sites are located within 50 bp of well-vetted MosSCI insertion sites on Chr II and Chr IV.
Description
Transgenic animals are powerful tools in the study of basic biological processes using C. elegans. The recent development of recombination-mediated cassette exchange (RMCE) integration approaches in worms provides a relatively rapid efficient method to create single copy transgenes with inserts up to at least 12 kb (Nonet, 2020,Yang et al., 2021). RMCE is similar to MosSCI (Frokjaer-Jensen et al., 2008) in that it depends on integrating at landing sites that have been engineered. In the case of MosSCI, a transposon is excised, and the double stranded break is repaired by a template often using synthesis-dependent strand annealing (SDSA) which is error prone (Frokjaer-Jensen et al., 2008). Similar approaches mediated by cas9 cleavage also have high error rates, approaching 65% in some studies (Au et al., 2019). By contrast, RMCE uses recombination to insert the template into the genome which rarely yield erroneous inserts (Nonet, 2020).
The RMCE approach I developed takes advantage of two distinct recombinases. A plasmid template delivered into the gonad of young adult animals is first integrated into the genome using FLP recombinase. Both the template and the landing site contain two distinct FLP integration sites, FRT and FRT3. Recombination between the FRT and FRT3 sites in the plasmid and the landing site yields replacement of the genomic FRT FRT3 interval with plasmid FRT FRT3 interval. This recombination likely occurs in two steps; first a loop in by recombination at one of the sites, followed by excision by recombination at the other. These recombination events typically occur in the F1 germline, though occasionally it occurs in the P0 animal (Nonet, 2020).
Two methods have been developed for performing RMCE. The first method uses a landing site and an unlinked source of FLP recombinase (usually bqSi711). Injection of the plasmid leads to integration of the plasmid at the landing site which is identified as a Rol (or HygR) animal. After the initial insertion is made homozygous, the self-excising marker cassette (SEC) is then excised using a heat shock Cre protocol, leading to the final insertion. The insertion is then outcrossed from bqSi711 using simple crosses. The second method utilizes a landing site which contains a germline FLP expressing transcription unit contained within the landing site. In this case, the FLP expression element is excised using a heat shock Cre protocol. One limitation of the RMCE approach is the lack of landing sites for integration. Here, I describe several new landing sites created using a CRISPR integration approach.
I integrated a two-component landing site using CRISPR/cas9 just adjacent to the position of the ttTi5605 Mos1 insertion that has been widely used for creating single copy insertions using MosSCI (Frokjaer-Jensen et al., 2008). I also integrated a single-component landing site at same position on Chr II and another at a site adjacent to cxTi10882, another commonly used Mos1 insertion site on Chr IV. In addition, I modified the previously described two-component landing site jsTi1453 I, deleting the left miniMosarm from that landing site (Fig. 1A, B). I first characterized these novel landing sites by integrating the identical mec-4p GFP-C1 tbb-2 3’ construct at each site and comparing the expression level of insertions at the new landing sites to identical insertions at the previously described landing sites (Nonet, 2020). The expression of GFP-C1 in touch receptor neurons (TRN) was easily detected in all the new landing site transgenes, though the expression level was slightly lower than that observed in integrations at the four previously described sites (Fig. 1C).
Recently developed bipartite reporters including a tetO/tetR Tet OFF system exhibit background expression in both the pharynx and the rectal gland cells (Nonet, 2020). Because a similar rectal gland background signal was observed at several different landing sites using distinct bipartite systems, I speculated that the miniMos transposon arm might be contributing to the background. Comparison of the identical tetO 7X ∆mec-7p GFP-C1 reporter integrated at both jsTi1453 and the js1570 ∆mosL arm derivative confirmed this was the case as expression in the rectal gland cells was undetectable in the js1570 derived transgene (Fig 1D). Despite the reduction in background, the tetO reporter still robustly expressed GFP in TRNs, when driven by the identical mec-4 promoter tet OFF driver (Fig. 1C).
In developing a new recombination-mediated homolog exchange technique (https://sites.wustl.edu/nonetlab/rmhe/), I have used these new landing sites to create addition RMCE insertions. I collated the insertion frequency data from a set of over 90 injection sessions in which I counted the number of Roller F1 animals obtained, and the number of insertions obtained (Fig. 1E). These data demonstrate that js1570, jsSi1579 and jsSi1669 all behave comparably to previously described landing sites, yielding insertions at a rate of approximately 1 per 3 injected animals. However, the jsSi1691 single component site yielded insertions at a lower frequency (1 per 10 injected animals). I speculate this is due to lower expression of FLP from the jsSi1691 landing site since integration at the same position using jsSi1579 and bqSi711 as a source of FLP yields normal integration frequencies. A recent study indicated that for CRISPR/cas9 genome modifications one could obtain a much higher frequency of integration events using specifically treated miniprep DNA (Huang et al., 2021). A preliminary set of 7 injections performed while this manuscript was under review and presented in Figure 1E suggest that similar benefits are seen for RMCE, raising the insertion frequency to above 1 integration per P0 in this admittedly small sample size.
I previously demonstrated that RMCE yields insertions of expected structure in greater than 95% of cases (Nonet, 2020). I have also characterized most of the transgenes obtained at these additional landing sites either by confirming the presence of an expected fluorescence pattern, recombinase activity, tetO reporter activity, or tetR driver activity. In some cases, long range PCR combined with restriction digests and/or sequencing was also performed to confirm the structure of insertions. All of the insertions at js1570 and jsSi1669 have been verified. The analysis of insertions on Chr II is ongoing, but to date only one of over 50 well-characterized insertions is incorrect with that insertion containing a distinct region of Chr II inserted adjacent to the loxP site. Three other cases of unexpected outcomes are also worth mentioning. In one case, I obtained a homozygous Rol insertion which still expressed the rpl-28p GFP-his-58 marker from the landing cassette. In addition, in two cases, I was unable to excise the SEC by heat shock. However, in all three cases, an independent sister insertion from the same injection session was used for the excision step to isolate the final insertion. I have not attempted to determine the molecular structure of these unusual ‘faulty’ insertions since they are easily identified and discarded.
The new landing site are now available at the CGC and should provide additional flexibility in creating RMCE-based transgenic animals. I also plan to create additional landing sites at well-characterized high expressing genomic positions on the remaining chromosomes that currently do not contain landing sites.
Methods
Request a detailed protocolC. elegans was maintained on NGM agar plates spotted with OP50 at 22.5°C or at 25°C during the RMCE protocol.
RMCE transgenesis
Inserts were cloned into pLF3FShC (Nonet, 2020), pRMHEB or pRMHEP (https://sites.wustl.edu/nonetlab/rmhe-vectors/) and injected at ~50 ng/µl into young adults. In the set of injections summarized in Figure 1E Qiagen miniprep DNA was prepared using a PB wash as described by Huang et al. (2021) and injected at 40-50 ng/ul. Integrants were identified and isolated as described in detail in Nonet (2020). Performing RMCE at 25°C is critical to obtaining robust integration rates. The criterion for including an injection session in the table (Fig. 1E) was obtaining a mean of at least 1 F1 Rol per injected animal. All injection sessions into js1570, jsSi1669 and jsSi1691 met this criterion. Eleven jsSi1579 injection sessions failed to meet this criterion. They consisted of 5 sessions injecting plasmids with strong ubiquitous promoters (eft-3 or rpl-27) driving tet OFF and 6 sessions injecting plasmids that contained both a tet OFF driver and a tetO reporter cassette. In all cases dead eggs were observed on the injection plates. In cases where a fluorescent protein reporter was in the plasmid, the dead eggs were brightly fluorescent. The two jsSi1579 failures included in the table were one rpl-27 session and one dual driver and reporter plasmid session. In some cases, I was able to integrate plasmids containing both a driver and a reporter by injecting animals growing on doxycycline.
CRISPR/cas9-mediated insertions and deletion
js1570 was created using a dpy-10 co-CRISPR strategy. A mix of plasmids NMp3143 (40 ng/ul), NMp3153 (10 ng/ul), NMp3828 (20 ng/ul), NMp3829 (20 ng/ul) and oligonucleotides NMo5238 (0.4 uM) and NMo6761 (0.4 uM) we co-injected into jsTi1453; bqSi711 animals. Rol progeny were screen by PCR (NMo6564/6569) for presence of the deletion and homozygosed. The deletion structure was confirmed by sequence analysis. jsSi1579 was created by injecting unc-119(ed3); bqSi711 animals with a mix of NMp3143 (40ng/ul), NMp3630 (50 ng/ul), NM3631 (30 ng/ul). Insertions were identified by selection for hygR Rol progeny by adding 25 ul of 100 mg/ml HydroGold™(InvivoGen) to P0 injection plates (3 animals per plate) 3 days after injection. After isolating homozygotes, the hygR sqt-1 self-excision cassette (SEC) in the insertion was excised by screening for non-Rol progeny after a 20-hr. heat shock at 29° C. The structure of the insertion was validated by a combination of long-range PCR [performed as outlined in Nonet (2020) using MNo3887/3888], restriction digestion and sequence analysis. jsSi1691 was created by injection of N2 (the wild type) with a mixture of plasmids NMp3143 (40 ng/ul), NMp3630 (50 ng/ul), NMp4043 (20 ng/ul), pBluescript KS (50 ng/ul), pGH8 (2 ng/ul) and pCFJ90 (2.5 ng/ul). Insertions were selected for and analyzed as described for jsSi1579. jsSi1669 was created by injection of oxTi1127 animals with a mixture of plasmid NM4055 (35 ng/ul), NM4057 (25 ng/ul), pBluescript KS (+) (50 ng/ul), pGH8 (2 ng/ul) and pCFJ90 (2.5 ng/ul). Insertions were identified by hygromycin selection as described above. The structure of the insertion was validated as described above using oligonucleotides NMo3889/3890. The genomic sequence of the insertions is available at https://sites.wustl.edu/nonetlab/rmce-insertion-strains/ .
Microscopy
For quantification of GFP signals, homozygous L4 hermaphrodite animals were mounted on 2% agar pads in a 2 µl drop of 1mM levamisole in phosphate buffered saline, cover slipped and imaged on an Olympus (Center Valley, PA) BX-60 microscope equipped with a Qimaging (Surrey, BC Canada) Retiga EXi monochrome CCD camera, a Lumencor AURA LED light source, Semrock (Rochester, NY) GFP-3035B and mCherry-A-000 filter sets, and a Tofra (Palo Alto, CA) focus drive, run using micro-manager 2.0ß software (Schindelin et al., 2012) using a 40X air lens at 20% LED power with 100 ms exposures. PLM soma and ALM soma signals were quantified using the FIJI version of ImageJ software (Edelstein et al., 2014) as described in Nonet (2020). Rectal gland cell images presented were collected using the same conditions.
Plasmid constructions
Integration vectors were assembled using Golden Gate (GG) reactions as described in Nonet (2020). Other plasmids were constructed using standard cloning techniques.
The following previously published plasmids were used:
pBluescript KS (+) (Short et al., 1988), pDD162 (Dickinson et al., 2013), pCFJ90 and pGH8 (Frokjaer-Jensen et al., 2008), and NMp3055, NMp3421, NMp3467, NMp3470, NMp3631, NMp3643, NMp3732, NMp3746 and NMp3774 (Nonet, 2020).
The following plasmids were constructed:
NMp3143 peft-3-cas9 (3 int)
A derivative of the pDD162 Cas9 expression plasmid lacking the empty U6 sgRNA cassette. pDD162 was amplified using NMo5228/5379 and re-circularized using a Gibson assembly reaction.
NMp3150 DR274 U6 FE
U6 promoter sgRNA clone with a flipped and extend sgRNA as described in Ward (2015). sgFE was amplified from NMp3055 using NMo5407/5075, purified, digested with BamHI and HindIII, and inserted into BamHI and HindIII digested NMp3055.
NMp3153 DR274 U6 FE dpy-10
U6 promoter sgRNA targeting dpy-10 at GCTACCATAGGCACCACgAG. NMp3150 was digested with BsaI and the annealed oligonucleotide pair NMo5236/5237 was inserted by ligation.
NMp3630 DR274 U6 MosII
U6 promoter sgRNA targeting Chr II adjacent to the ttTi5605 Mos1 insertion site at gatatcagtctgtttcgtaa cgg. NMp3055 was digested with BsaI and the annealed oligonucleotide pair NMo6439/6440 was inserted by ligation.
NMp3828 DR274 U6 sgTi1453 F
U6 promoter sgRNA targeting Chr I adjacent to the jsTi1453 landing site at attcacggcacaacatacat tgg. NMp3055 was digested with BsaI and the annealed oligonucleotide pair NMo6757/6758 was inserted by ligation.
NMp3829 DR274 U6 sgMosL
U6 promoter sgRNA targeting miniMos adjacent to the left arm at gttgAGCTCCACCGCGGTGG CGG. NMp3055 was digested with BsaI and the annealed oligonucleotide pair NMo6759/6760 was inserted by ligation.
NMp4043 pSAP ChrII FLP loxP FRT FRT3 landing
Chr II full RMCE landing site CRISPR template. The left arm amplified from N2 genomic DNA using NMo6707/6450, the right arm amplified from N2 genomic DNA using NM06708/6453, and the loxP FLP FRT FRT landing site from NMp3746 were co assembled into NMp3421 using a SapI Golden Gate assembly reaction.
NMp4053 DR274 5′ arm cxTi10882 left arm
Left arm genomic fragment adjacent to cxTi10882. The left arm was amplified from N2 genomic DNA using NMo7060/7061 and inserted into NMp3467 using a BsaI Golden Gate reaction.
NMp4054 DR274 3′ arm cxTi10882 right arm
Right arm genomic fragment adjacent to cxTi10882. The right arm was amplified from N2 genomic DNA using NMo7058/7059 and inserted into NMp3470 using a BsaI Golden Gate reaction.
NMp4055 DR274 U6 cxTi10882
U6 promoter sgRNA targeting Chr IV adjacent to the cxTi10882 Mos1 insertion site at actgttggatgcctgtgtag cgg. NMp3055 was digested with BsaI and the annealed oligonucleotide pair NMo7062/7063 was inserted by ligation.
NMp4057 pSAP cxTi10882 FLP loxP FRT FRT3 landing
Chr IV full RMCE landing site CRISPR template. The left chr IV arm from NMp4053, the right Chr IV arm from NMp4054 and the loxP FLP FRT FRT landing site from NMp3746 were co assembled into NMp3421 using a SapI Golden Gate assembly reaction.
Oligonucleotides
| NMo number | Sequence 5’ > 3’ |
| 3887 | ACCGGAAACCAAAGGACGAGAG |
| 3888 | ACGCCCAGGAGAACACGTTAG |
| 3889 | CCAAACAAGTGTCGTTGACCCAG |
| 3890 | CATATCCGCCAAGGACGCTC |
| 5075 | GCCAAGCTTCACAGCCGACTATGTTTGGCGTC |
| 5228 | CGCCAGGGTTTTCCCAGTCACGACGTTGTAAAACGACGGCCAGTG |
| 5236 | TTTGCTACCATAGGCACCACGAG |
| 5237 | AAACCTCGTGGTGCCTATGGTAG |
| 5238 | ACTTGAACTTCAATACGGCAAGATGAGAATGACTGGAAACCGTACCGCATGCGGTGCCTATGGTAGCGGAGCTTCACATGGCTTCAGACCAACAGCCTAT |
| 5379 | CGTCGTGACTGGGAAAACCCTGGCGTTCCCAACAGTTGCGCAGCC |
| 5407 | AAGGATCCGGGTCTCAGTTTAAGAGCTATGCTGGAAACAGCATAGCAAGTTTAAATAAGGCTAGTCCG |
| 6439 | TTTGATATCAGTCTGTTTCGTAA |
| 6440 | AAACTTACGAAACAGACTGATAT |
| 6450 | TTGCTCTTCATGGCCTCTGAACTGGTACCTC |
| 6453 | TTGCTCTTCATACCTTGCCATTGTTTCCTG |
| 6564 | CATCCCATTCACGGCACAAC |
| 6569 | AAGCTCTTCACTCCGCATTTTCTCCCACCCTG |
| 6707 | AAGCTCTTCACGCGAAACAGACTGATATCGAAAC |
| 6708 | AAGCTCTTCAACGTAACGGTCTTCTGTATAAC |
| 6757 | TTTGATTCACGGCACAACATACAT |
| 6758 | AAACATGTATGTTGTGCCGTGAAT |
| 6759 | TTTGATTCACGGCACAACATACAT |
| 6760 | AAACATGTATGTTGTGCCGTGAAT |
| 6761 | AACAATTCATCCCATTCACGGCACAACATATGGCGGCCGCTCTAGAACTAGGCTGTTTCG |
| 7058 | AGGTCTCAGACGCTGTGTAGCGGTCCTCTATTG |
| 7059 | AGGTCTCTCTACAGTCGCATACGTCGTATCCC |
| 7060 | GGTCTCAGTGGTTTCAACGGTGGAAGAAGGG |
| 7061 | CGGTCTCTTCGCACAGGCATCCAACAGTACG |
| 7062 | TTTGACTGTTGGATGCCTGTGTAG |
| 7063 | AAACCTACACAGGCATCCAACAGT |
Novel transgenes
| Transgene | Description | Full Designation | Comments |
| js1570 I | Chr I landing site | jsTi1453 js1570 [∆mosL loxP rpl-28p FRT GFP-his-58 FRT3 mosR] I | This study |
| jsSi1579 II | Chr II landing site | jsSi1579 [loxP rpl-28p FRT GFP-his-58 FRT3] II | This study |
| jsSi1669 IV | Chr IV landing site | jsTi1669 [loxP mex-5p FLP D5 sl2 mNG glh-2 3′ rpl-28p FRT GFP-his-58 FRT3] IV | This study |
| jsSi1691 II | Chr II landing site | jsTi1691 [loxP mex-5p FLP D5 sl2 mNG glh-2 3′ rpl-28p FRT GFP-his-58 FRT3] II | This study |
| jsSi1571 I | 7X tetO GFP-C1 | jsTi1453 js1570 jsSi1571 [∆mosL loxP tetO 7X ∆mec-7p GFP-C1 tbb-2 3′ FRT3 mosR] I | This study. RMCE insertion of NMp3774 into js1570 |
| jsSi1684 IV | mec-4p GFP-C1 | jsTi1669 jsSi1684 [loxP mec-4p GFP-C1 tbb-2 3′ FRT3] IV | This study. RMCE insertion of NMp3732 into jsSi1669 |
| jsSi1707 II | mec-4p GFP-C1 | jsSi1579 jsSi1707 [loxP mec-4p GFP-C1 tbb-2 3′ FRT3] II | This study. RMCE insertion of NMp3732 into jsSi1579 |
| jsSi1734 I | mec-4p GFP-C1 | jsTi1453 js1570 jsSi1734 [∆mosL loxP mec-4p GFP-C1 tbb-2 3′ FRT3 mosR] I | This study. RMCE insertion of NMp3732 into js1570 |
Worm Strains
| Strain | Genotype | Source |
| BN711 | unc-119(ed3) III; bqSi711 [mex-5p::FLP::SL2::mNG + unc-119(+)] IV | Macías-León and
Askjaer (2018); CGC |
| EG8992 | F53A2.9(oxTi1127[Pmex-5::Cas9::tbb-2 3’UTR, Phsp-16.41::Cre::tbb-2 3’UTR, Pmyo-2::nls-CyOFP::let-858 3’UTR + lox2272]) III | Schwartz et al. (2021); CGC |
| NM5196 | jsTi1493 jsSi1502 [mosL loxP mec-4p GFP-C1 tbb-2 3′ FRT3 mosR] IV | Nonet (2020); CGC |
| NM5209 | jsTi1453 jsSi1514 [mosL loxP mec-4p GFP-C1 tbb-2 3′ FRT3 mosR] I; him-8(e1489) IV | Nonet (2020) |
| NM5228 | jsTi1490 jsSi1529 [mosL loxP mec-4p GFP-C1 tbb-2 3′ FRT3 mosR] IV | Nonet (2020); CGC |
| NM5236 | jsTi1492 jsSi1535 [mosL loxP mec-4p GFP-C1 tbb-2 3′ FRT3 mosR] II | Nonet (2020) |
| NM5264 | jsTi1453 jsSi1543 [mosL loxP tetO 7X ∆mec-7p GFP-C1 tbb-2 3′ FRT3 mosR] I; him-8(e1489) IV | Nonet (2020) |
| NM5295 | jsTi1493 jsSi1560 [mosL loxP mec-4p tetR-L-QFADact-4 3′ FRT3 mosR] IV | Nonet (2020) |
| NM5322 | jsTi1453 js1570 [∆mosL loxP rpl-28p FRT GFP-his-58 FRT3 mosR] I; bqSi711 IV | This study |
| NM5327 | jsSi1453 js1570 jsSi1571[loxP tetO 7X ∆mec-7p GFP-C1 tbb-2 3′ FRT3 mosR] I; him-8(e1489) IV | This study |
| NM5337 | jsSi1453 js1570 jsSi1571 [loxP tetO 7X ∆mec-7p GFP-C1 tbb-2 3′ FRT3 mosR] I; jsTi1493 jsSi1560 [mosL loxP mec-4p tetR-L-QFAD act-4 3′ FRT3 mosR] IV | This study |
| NM5402 | jsSi1579 [loxP rpl-28p FRT GFP-his-58 FRT3] II; bqSi711 IV | This study |
| NM5467 | jsTi1453 jsSi1543 [mosL loxP tetO 7X ∆mec-7p GFP-C1 tbb-2 3′ FRT3 mosR] I; jsTi1493 jsSi1560 [mosL loxP mec-4p tetR-L-QFAD act-4 3’ FRT3 mosR] IV | Nonet (2021) |
| NM5471 | jsTi1669 [loxP mex-5p FLP D5 sl2 mNG glh-2 3′ rpl-28p FRT GFP-his-58 FRT3] IV | This study |
| NM5500 | jsTi1691 [loxP mex-5p FLP D5 sl2 mNG glh-2 3′ rpl-28p FRT GFP-his-58 FRT3] II | This study |
| NM5580 | jsTi1579 jsSi1707 [loxP mec-4p GFP-C1 tbb-2 3′ FRT3] II | This study |
| NM5582 | jsTi1669 jsSi1684 [loxP mec-4p GFP-C1 tbb-2 3′ FRT3] IV | This study |
| NM5633 | jsTi1453 js1570 jsSi1734 [∆mosL loxP mec-4p GFP-C1 tbb-2 3′ FRT3 mosR] I; him-8(e1489) IV | This study |
Reagents
Plasmids are available by request from MLN. Strains containing the four new landing sites have been submitted to the Caenorhabditis Genetics Center (CGC). Plasmids and additional strains and will be submitted to Addgene or CGC and if demand levels warrant it.
References
Funding
WUMS Department of Neuroscience funds and R01 GM141688.
Reviewed By
Christian Frøkjær-JensenHistory
Received: October 27, 2021Revision received: December 8, 2021
Accepted: December 15, 2021
Published: December 16, 2021
Copyright
© 2021 by the authors. This is an open-access article distributed under the terms of the Creative Commons Attribution 4.0 International (CC BY 4.0) License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.Citation
Nonet, M (2021). Additional Landing Sites for Recombination-Mediated Cassette Exchange in C. elegans. microPublication Biology. 10.17912/micropub.biology.000503.Download: RIS BibTeX