Nanobiology Institute, Yale University
Department of Cell Biology, Yale University School of Medicine
Abstract
Clathrin mediated endocytosis (CME) in the fission yeast Schizosaccharomyces pombe critically depends on the connection between the lipid membrane and F-actin. The fission yeast endocytic protein End4 (homologous to Sla2 in budding yeast and HIP1R in human) contains a N-terminal domain that binds to PIP2 on the membrane, and a C-terminal THATCH domain that is postulated to be a binding partner of F-actin in vivo. Purified THATCH domain of the budding yeast Sla2, however, shows low affinity to F-actin in vitro. We tested if isolated THATCH domain still has low affinity to F-actin in vivo, using TEV protease (TEVp)-mediated protein cleaving to separate the THATCH domain from the rest of End4. Our results indicate that the isolated THATCH domain of End4 is unable to bind F-actin independently in vivo, consistent with the low affinity of the THATCH domain to F-actin measured from in vitro binding assays.
Description
End4 is an endocytic protein that mediates the connection between the lipid membrane and the actin cytoskeleton in fission yeast (Gottfried, Ehrlich, and Ashery 2010, 4; Iwaki et al. 2004, 4; Lacy et al. 2018, 4; Engqvist-Goldstein et al. 2001; Skruzny et al. 2012). The N-terminus of End4 binds to PIP2 on the membrane together with Ent1p, and the C-terminus of End4 shares sequence similarities to the actin binding domain of talin, and is therefore named THATCH (talin-HIP1/R/Sla2p actin-tethering C-terminal homology) (Legendre-Guillemin 2004, 4; Baggett, D’Aquino, and Wendland 2003, 4; McCann and Craig 1997; Brett et al. 2006). The crystal structures of THATCH domains from HIP1R and talin are five-helix-bundles (Brett et al. 2006; Gingras et al. 2008). The THATCH domain of Sla2 is critical for linking F-actin to the endocytic coat during CME in budding yeast (Baggett, D’Aquino, and Wendland 2003, 2; Brett et al. 2006; Abella et al. 2021). The affinity of purified THATCH domains for actin filaments has been measured in vitro by multiple groups (Brett et al. 2006; Gingras et al. 2008; Senetar, Foster, and McCann 2004). Although structurally similar, the THATCH domain from HIP1R has the lowest affinity to actin among all tested THATCH domains, in some cases not significantly higher than that of the negative control with purified GST (Senetar, Foster, and McCann 2004). This discrepancy in the actin-binding ability of the THATCH domain from in vitro and in vivo data could be explained by cryptic actin-binding sites that are activated in vivo, by a) post-translational modifications, b) the binding of other molecules, or c) force transmitted through the rest of the End4 molecule. All three binding site activation mechanisms have been found on talin, the molecule responsible for connecting the membrane to F-actin during cell adhesion through its 13 rod domains including the THATCH domain (Goult, Yan, and Schwartz 2018; Goult, Brown, and Schwartz 2021). A clear way to distinguish mechanism c) from a) and b) is through in vivo protein cleaving. By isolating the THATCH domain of End4 from the rest of End4 in the cytoplasm of the fission yeast, the native environment for possible post-translational modifications or binding partners of THATCH are preserved, but force transmission to the THATCH domain is prevented. Consequently, the cellular localization of the isolated THATCH domain is indicative of its direct affinity to F-actin in vivo.
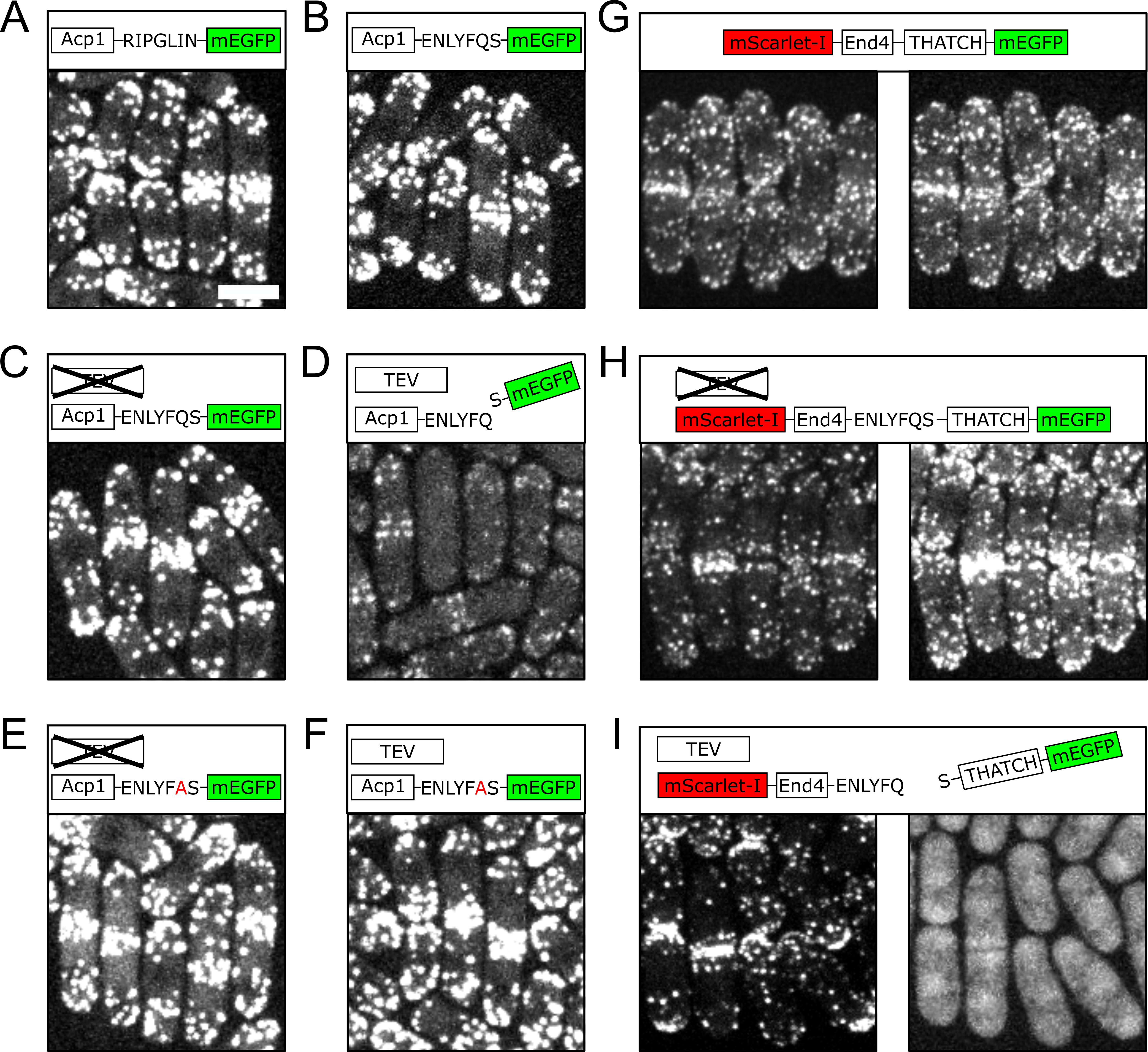
We chose to use TEVp to perform in vivo protein cleaving, because TEVp has good specificity and only needs seven amino acids as the recognition sequence (Sanchez and Ting 2020). TEVp has been shown to work in diverse situations both in vivo and in vitro (Harder et al. 2008; Raran-Kurussi et al. 2017). The coding sequence of TEVp was integrated into the genome of fission yeast by CRISPR/Cas9(Fernandez and Berro 2016), and put under the control of the inducible promoter nmt1 (Forsburg 1993). We first verified that our approach for in vivo protein cleaving worked and did not have undesirable effects, by using TEVp to separate the mEGFP tag from a control protein. We used the Acp1 subunit of the canonical actin filament capping protein, which localizes to endocytic sites (Sun et al. 2019; Nakano and Mabuchi 2006; Berro and Pollard 2014) and has comparable cytoplasmic concentration to End4 in the fission yeast (Sirotkin et al. 2010). When Acp1 was tagged either with a regular linker or the TEVp recognition peptide, the majority of the mEGFP signal was detected at endocytic patches, which were enriched at the poles and the division plane of the fission yeast cells (Fig. 1A-C). These endocytic patches contained a large amount of F-actin(Lacy et al. 2018). When the expression of TEVp was induced, we saw a drastic decrease of mEGFP signal from endocytic patches, suggesting the removal of the mEGFP tag from Acp1 molecules (Fig. 1D). As expected, the cleavage of TEVp demonstrated good specificity, and a point mutation in the TEVp recognition peptide prevented protein cleaving (Fig. 1E, 1F).
We then inserted the TEVp recognition peptide prior to the THATCH domain of End4. In wild-type cells, End4 localizes to endocytic patches in a pattern similar to Acp1 (Fig. 1G). The insertion of the TEVp recognition peptide did not change the localization of End4 when TEVp was not expressed (Fig. 1H). When TEVp was expressed, however, the THATCH domain was no longer found at endocytic patches but became diffusive in the cytoplasm and the nucleus, while the N-terminal fragment of End4 still showed polarized localization (Fig. 1I). Failure of the isolated THACTH domain to localize to endocytic patches demonstrates that the affinity of the THATCH domain to F-actin is low in vivo, similarly to what was shown in vitro. In addition, our results demonstrate that the connection between the THATCH domain and the rest of the End4 molecule is necessary for its binding to actin filaments. We speculate that force transmission through the whole End4 protein is probably involved in exposing the cryptic F-actin binding sites.
Methods
Request a detailed protocolYeast strains and media
The S. pombe strains used in this study were made though CRISPR/Cas9 mediated genome editing as reported in (Fernandez and Berro 2016), and the edited gene sequences were confirmed by colony PCR and sequencing. The coding sequence of TEVp was inserted into the low complex region 0.5kb upstream of pil1 locus (chromosome III, insertion after 348203) (V. Wood et al. 2002; Valerie Wood et al. 2012), under the control of the full strength nmt1 promoter (Forsburg 1993). S. pombe cells were cultured in YE5S (Yeast Extract supplemented with 0.225 g/L of uracil, lysine, histidine, adenine and leucine), and imaged in EMM5S (Edinburgh Minimum media supplemented with 0.225 g/L of uracil, histidine, adenine, lysine, and leucine) after washing with EMM5S. Yeast cells were cultured at 32 °C with 200rpm shaking overnight to reach OD595nm reading between 0.3 and 0.5. The expression of TEVp was inhibited when yeast cells were cultured in YE5S media. The expression of TEVp was induced by transferring yeast cells into EMM5S media after multiple washes, and cells were imaged after overnight culture in EMM5S to allow for enough time for TEVp mediated protein cleavage.
Microscopy
Cells were imaged at room temperature on gelatin pads (25%) on glass slides. Samples were mounted on a Nikon TiE inverted microscope (Nikon, Tokyo, Japan) with a CSU-W1 Confocal Scanning Unit (Yokogawa Electric Corporation, Tokyo, Japan) with a CFI Plan Apo 100X/1.45NA Phase objective (Nikon, Tokyo, Japan). Images were captured with an iXon Ultra888 EMCCD camera (Andor, Belfast, UK). mEGFP tagged strains were excited with a 488-nm argon-ion laser and filtered with a single band pass filter 510/25. mScarlet-I tagged strains were excited with a 561-nm argon-ion laser and filtered with a single band pass filter 575/25. Fluorescent signals from the whole cell were collected by taking 21 optical sections each with 0.5µm thickness, and 2D maximum projected images were created for each strain by the Fiji distribution of ImageJ (Schindelin et al. 2012). All Acp1 strains were imaged and displayed with the same settings. For End4 strains, the mEGFP channel was imaged and displayed with the same settings, and the mScarlet-I channel was imaged and displayed with the same settings.
Reagents
| Name | Genotype | Used in | Source |
| SpJB366 | acp1-mEGFP fex1Δ fex2Δ ade6-M216 his3-D1 leu1-32 ura4-D18 h- | A | This Study |
| SpJB524 | acp1-TEVSite-mEGFP fex1Δ fex2Δ ade6-M216 his3-D1 leu1-32 ura4-D18 h- | B | This Study |
| SpJB535 | nmt-TEVp acp1-TEVSite-mEGFP fex1Δ fex2Δ ade6-M216 his3-D1 leu1-32 ura4-D18 h- | C, D | This Study |
| SpJB561 | mScarlet-I-end4-mEGFP fex1Δ fex2Δ ade6-M216 his3-D1 leu1-32 ura4-D18 h- | G | (Ren et al. 2021) |
| SpJB572 | nmt-TEVp mScarlet-I-end4-TEVSite-end4-mEGFP fex1Δ fex2Δ ade6-M216 his3-D1 leu1-32 ura4-D18 h- | H, I | This Study |
| SpJB583 | nmt-TEVp acp1-TEVSite_QP1A-mEGFP fex1Δ fex2Δ ade6-M216 his3-D1 leu1-32 ura4-D18 h- | E, F | This Study |
Acknowledgments
We thank members of the Berro lab for helpful discussions and insights, especially Ronan Fernandez for suggesting important controls. We also thank the Yale West Campus Imaging Core for their spinning disc confocal microscope. We thank the Keck DNA Sequencing Facility at Yale for their assistance with oligo synthesis and sequencing.
References
Funding
This research was supported by National Institutes of Health/National Institute of General Medical Sciences Grant R21GM132661.
Reviewed By
AnonymousHistory
Received: November 30, 2021Revision received: January 3, 2022
Accepted: January 3, 2022
Published: January 6, 2022
Copyright
© 2022 by the authors. This is an open-access article distributed under the terms of the Creative Commons Attribution 4.0 International (CC BY 4.0) License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.Citation
Ren, Y; Berro, J (2022). Isolated THATCH domain of End4 is unable to bind F-actin independently in the fission yeast Schizosaccharomyces pombe. microPublication Biology. 10.17912/micropub.biology.000508.Download: RIS BibTeX