Center of Parasitology, Severtsov Institute of Ecology and Evolution RAS, Moscow, Russia
Division of Aging Biology, National Institute on Aging, Bethesda, MD, 20892, USA
The Buck Institute for Research on Aging, Novato, CA, 94945, USA
Rutgers University, Dept. of Molecular Biology and Biochemistry, Piscataway, NJ, 08854, USA
Abstract
The Caenorhabditis Intervention Testing Program (CITP) was founded on the principle that compounds with positive effects across a genetically diverse test-set should have an increased probability of engaging conserved biochemical pathways with mammalian translational potential. To fulfill its mandate, the CITP uses a genetic diversity panel of Caenorhabditis strains for assaying longevity effects of candidate compounds. The panel comprises 22 strains from three different species, collected globally, to achieve inter-population genetic diversity. The three represented species, C. elegans, C. briggsae, and C. tropicalis, are all sequential hermaphrodites, which simplifies experimental procedures while maximizing intra-population homogeneity. Here, we present estimates of the genetic diversity encapsulated by the constituent strains in the panel based on their most recently published and publicly available whole-genome sequences, as well as two newly generated genomic data sets. We observed average genome-wide nucleotide diversity (π) within the C. elegans (1.2e-3), C. briggsae (7.5e-3), and C. tropicalis strains (2.6e-3) greater than estimates for human populations, and comparable to that found in mouse populations. Our analysis supports the assumption that the CITP screening panel encompasses broad genetic diversity, suggesting that lifespan-extending chemicals with efficacy across the panel should be enriched for interventions that function on conserved processes that are shared across genetic backgrounds. While the diversity panel was established by the CITP for studying longevity interventions, the panel may prove useful for the broader research community when seeking broadly efficacious interventions for any phenotype with potential genetic background effects.
Description
Model organisms have been fruitful tools for elucidating core biological principles. The power of model organism study, in part, is due to the ability to grow large populations with known genetic makeup. One of the most widely adopted genetic models is the hermaphroditic nematode Caenorhabditis elegans. The reproductive style of C. elegans makes it particularly easy to generate and maintain large populations of genetically identical individuals. In fact, the control over genetic variability helped make C. elegans the first multi-cellular organism to have its entire genome sequenced (C. elegans Sequencing Consortium 1998). Despite the success garnered using genetically homogeneous populations, it has become increasingly apparent that many of the phenotypes of interest for study are dependent on genetic background (see Evans et al. 2021 for review). Examples of these background-influenced phenotypes range from α-synuclein toxicity (Wang et al. 2019), to behavioral responses to temperature (Stegeman et al. 2013), dietary influence on lifespan and reproduction (Stastna et al. 2015), and pharmacological efficacy (Lucanic et al. 2017). The dependence on genetic background suggests that attempts to identify core biological systems and functionality could benefit from assaying across genetic diversity to identify genetic background-independent phenotypes. One way to achieve this is to use a panel of populations with intra-population homogeneity and inter-population diversity.
With the importance of genetic background effects in mind, the Caenorhabditis Intervention Testing Program (CITP) was designed to identify anti-aging and longevity-promoting compound interventions effective in a genetically diverse set of Caenorhabditis nematode populations (Lucanic et al. 2017). To date, 55 chemical compounds have been tested for reproducible, genetic background-independent effects on longevity (Lucanic et al. 2017; Banse et al. 2019; Coleman-Hulbert et al. 2019; Coleman-Hulbert et al. 2020; Morshead et al. 2020; Osman et al. 2021; Banse et al. 2021; Onken et al. 2021). The genetic diversity panel used by the CITP is composed of 22 strains from three hermaphroditic species, which facilitates maintenance of intra-population homogeneity. While the full panel is composed of 22 stains, a smaller core sub-panel of nine strains (three from each of three species, C. elegans (N2-PD1073, JU775, MY16), C. briggsae (AF16, HK104, ED3092), and C. tropicalis (JU1373, JU1630, QG843) is used in initial compound effect characterization (Banse et al. 2021). Here, we present estimates of the genetic diversity encapsulated by the constituent strains of the CITP panel, and core sub-panel, based on their most recently published and publicly available whole-genome sequences.
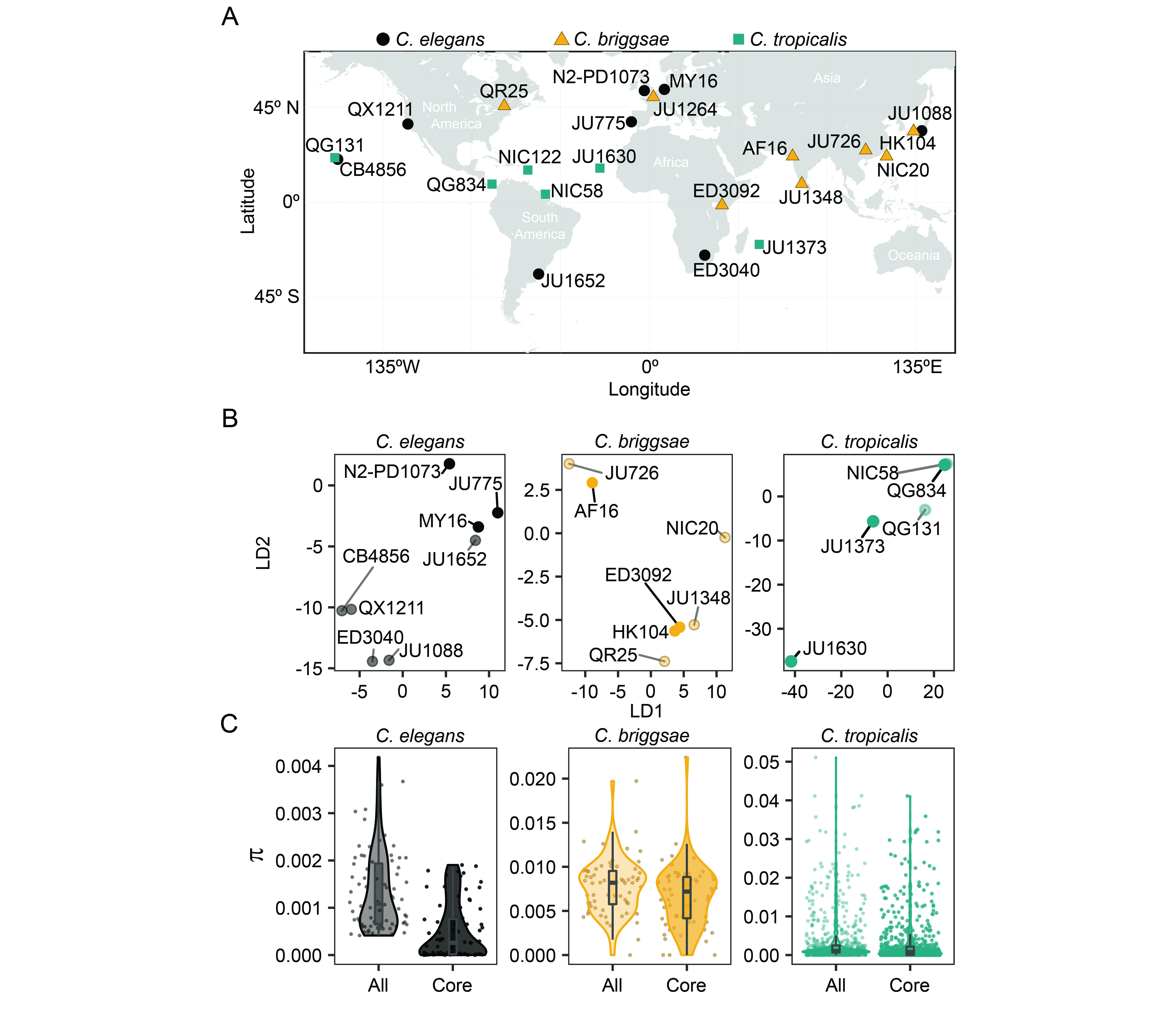
When establishing the genetic diversity panel for compound screening, the CITP sought strains that represented both broad geographic and genetic diversity. The three species represented in the panel are themselves globally distributed, but with ecological restrictions. For example, C. elegans is typically isolated from cooler ecological niches than C. tropicalis (Kiontke et al. 2011; Frézal and Félix 2015; Noble et al. 2021), while C. briggsae is found in niches that range from cool to warm (Frézal and Félix 2015). The wide species distributions enabled the CITP to assemble a panel of strains isolated worldwide, with representatives from most continents (Figure 1A). We next sought to determine the genetic diversity encapsulated by the panel by collecting genomic data for the strains (see Reagents) and analyzing the genomes for variation within each species (see Software). To visualize the population structure within the panel for each species we used a variational autoencoder approach, popVAE (Battey et al. 2021), to project genotypes of the strains on two latent dimensions (Figure 1B). We then determined the nucleotide diversity in the panel. The observed average genome-wide nucleotide diversity (π) among the C. elegans, C. briggsae, and C. tropicalis strains were, respectively, 1.2e-3, 7.5e-3, and 2.6e-3 (3.8e-4, 6.2e-3, and 2.4e-3 for the nine core CITP strains), which is consistent with previous estimates for those species (Graustein et al. 2002; Jovelin et al. 2009; Andersen et al. 2012; Crombie et al. 2019; Noble et al. 2021). Figure 1C shows nucleotide diversity estimated on 100 kb windows along the genomes for both the 20 strains in the full panel with available sequencing data, and for the nine strains in the core sub-panel. The estimated level of genetic diversity within these Caenorhabditis species is higher than that within human populations (Yu et al. 2004; Perry et al. 2013; Prado-Martinez et al. 2013; Arbiza et al. 2014; 1000 Genomes Project Consortium et al. 2015) and comparable to that found in mouse populations (Halligan et al. 2010; Booker and Keightley 2018). Our analysis supports the assumption that the CITP screening panel encompasses broad genetic diversity, suggesting that lifespan-extending chemicals with efficacy across the panel should be enriched for interventions that function on conserved processes that are shared across genetic backgrounds. While the diversity panel was established by the CITP for studying longevity interventions, the panel may prove useful for the broader research community when seeking broadly efficacious interventions for any phenotype with potential genetic background effects.
Methods
Request a detailed protocolTo generate estimates of the genetic variability among the strains in the CITP diversity panel we collected publicly available genomic data for 18 of the 22 CITP strains (see Reagents below). Because comparable Illumina-based sequencing was unavailable for N2-PD1073 and ED3092, two strains present in the core-subpanel of nine strains, we generated whole genome data for these two strains using standard protocols. In brief, we used proteinase K to digest whole worms and isolated total genomic DNA using the Genomic DNA Clean & Concentrator kit (Zymo Research). Genomic libraries were then generated using the Nextera XT DNA Library Preparation Kit (Illumina, Inc.). Sequencing was carried out on the Illumina NovaSeq 6000 platform by the Genomics and Cell Characterization Core Facility (GC3F) at the University of Oregon. We then used the two new genomic datasets, along with the 18 available in the NCBI database, to calculate genetic diversity estimates using our software pipeline (described below).
Reagents
While most of the strains in the CITP diversity panel can be obtained from the Caenorhabditis Genetics Center (CGC), the C. elegans wild-isolates should be ordered from the Caenorhabditis elegans Natural Diversity Resource (CeNDR) (Cook et al. 2017). Strains unavailable from those sources (e.g., N2-PD1073) can be obtained directly from the CITP upon request. Relevant to the unavailability of PD1073 at the CGC, PD1073 is a close relative of PD1074 which is distributed by the CGC as a wild type reference strain. PD1073, PD1074, and PD1075 are three subclones of the N2-derivative strain VC2010 that were generated in the process of assembling a new N2 reference (“VC2010-1.0”) genome (Yoshimura et al. 2019). All strains in the genetic diversity panel and the associated SRA read accession IDs used in this study are listed below:
C. elegans: strain CB4856 (with SRA read accession numbers SRR9322768, SRR9322769, SRR9322775, SRR9322863, SRR9322864), ED3040 (SRR9322720, SRR9322724, SRR9322725, SRR9322726, SRR9322818), JU1088 (SRR9322295, SRR9322300, SRR9322301), JU1652 (SRR9322577, SRR9322578, SRR9322579), JU775 (SRR9322349, SRR9322352, SRR9322355, SRR9322364), MY16 (SRR9322907, SRR9322327, SRR9322913, SRR9322161), QX1211 (SRR9324168, SRR9324217, SRR9323954, SRR9323955);
C. briggsae: AF16 (SRR2002620), HK104 (ERR3063412, ERR3063411, SRR8333803), JU1348 (SRR1793004), JU726 (SRR1792964), NIC20 (SRR1793012), QR25 (SRR1793006);
C. tropicalis: JU1373 (SRR12623131, SRR241785), JU1630 (SRR12623130), NIC58 (SRR12623045, SRR12623063), QG131 (SRR12623047), QG834 (SRR12623046).
No genomic data is publicly available for C. briggsae strain JU1264 and C. tropicalis NIC122.
Raw reads for C. elegans N2-PD1073 and C. briggsae ED3092 strains generated in this study were deposited to the SRA database (https://www.ncbi.nlm.nih.gov/sra) under the project accession ID PRJNA773598.
The core CITP strains are C. elegans N2-PD1073, MY16, and JU775; C. briggsae AF16, ED3092, and HK104; C. tropicalis JU1373, JU1630, and QG834.
Software
We estimated genetic diversity of the CITP strains using whole-genomic data for 20 out of 22 CITP strains from the panel. Reads from the SRA database were downloaded with SRA-toolkit (the SRA Toolkit Development Team). We evaluated the read quality with FastQC v.0.11.5 (Andrews 2010) and MultiQC v.1.3 (Ewels et al. 2016), and filtered and trimmed reads with Skewer v.0.2.2 (Jiang et al. 2014). We mapped filtered reads to the C. elegans, C. briggsae, and C. tropicalis genomes obtained from the WormBase database (https://wormbase.org//) version WS280 (with accession ID PRJNA13758, PRJNA10731, PRJNA53597, respectively) by BWA-MEM v.0.7.17 (Li 2013) and SAMtools v.1.5 (Li et al. 2009), and deduplicated with the Picard tools v.2.17.6 (Broad Institute). We called and filtered variants using GATK v.3.7 (McKenna et al. 2010) software and BEDtools v.2.25 (Quinlan and Hall 2010), and estimated the nucleotide diversity using VCFtools v.0.1.15 (Danecek et al. 2011), masking repetitive regions, indels, complex variants, and regions with too low or high coverage. We projected genotypes of the CITP strains on two latent dimensions using popVAE (Battey et al. 2021), and visualized them in R v.3.5 (R Core team 2018) using packages ggplot2 (Wickham 2016) and ggrepel.
All the tools used for the analysis are publicly available and free, the scripts used to get the results and figures are in the GitHub repository (https://github.com/phillips-lab/CITP_diversity/) and are available under the MIT license. The global map was generated in MATLAB R2021b (MathWorks, Inc.) using the MATLAB Mapping Toolbox. The location coordinates used for the strains are included in the GitHub repository. Figure readability was improved by moving panel relative positions, updating color coding, and improving text aesthetics using Adobe Illustrator 2022 in a manner consistent with image integrity standards.
Acknowledgments
We acknowledge the members of the Phillips labs for helpful discussions. We thank the CITP Advisory Committee, Max Guo and Ronald Kohanski (National Institute on Aging) for extensive discussion. This work was conducted using the resources of the Research Advanced Computing Services at the University of Oregon.
References
Funding
This work was supported by funding from National Institutes of Health grants (U01 AG045844, U01 AG045864, U01 AG045829, U24 AG056052, and R35 GM131838)
Reviewed By
Erik AndersenHistory
Received: November 22, 2021Revision received: December 15, 2021
Accepted: January 11, 2022
Published: January 27, 2022
Copyright
© 2022 by the authors. This is an open-access article distributed under the terms of the Creative Commons Attribution 4.0 International (CC BY 4.0) License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.Citation
Teterina, AA; Coleman-Hulbert, AL; Banse, SA; Willis, JH; Perez, VI; Lithgow, GJ; Driscoll, M; Phillips, PC (2022). Genetic diversity estimates for the Caenorhabditis Intervention Testing Program screening panel. microPublication Biology. 10.17912/micropub.biology.000518.Download: RIS BibTeX