Department of Integrative Biology, University of Wisconsin-Madison, USA
Abstract
Dorsal intercalation of the embryonic epidermis in the Caenorhabditis elegans embryo is a promising system for genetic analysis of convergent extension, a conserved process in animal embryos. We sought to identify functionally important actin regulators in dorsal epidermal cells. A promising candidate is MIG-10, the single MIG-10/RIAM/Lamellipodin (MRL) family member in C. elegans. We endogenously tagged all mig-10 isoforms with mNeonGreen and analyzed mig-10 mutants using 4-dimensional microscopy. MIG-10::mNG is expressed prominently in muscle progenitors but is not detectable in the dorsal epidermis. mig-10(ct41) homozygotes complete dorsal intercalation in a manner indistinguishable from wildtype, indicating MIG-10 is not essential during dorsal intercalation.
Description
Results and Discussion
Convergent extension, the mediolateral interdigitation of cells to elongate a tissue array along the anterior-posterior axis, is a conserved feature of embryonic development in animals, and underlies such key events in vertebrates as gastrulation and neurulation. In the Caenorhabditis elegans embryo a convergent extension-like movement known as dorsal intercalation occurs in the embryonic epidermis. The 20 dorsal epidermal cells are born as two rows of 10 cells each that interdigitate to form a single row that straddles the dorsal midline (Williams-Masson et al., 1998). This process, particularly within the posterior dorsal epidermis derived from the C blastomere, is a highly stereotypical process in wild-type embryos (Williams-Masson et al., 1998), making it a promising system for genetic analysis of a simple example of epithelial convergent extension. Dorsal intercalation requires extension of medially directed basolateral protrusions by intercalating cells regulated by Rho family GTPases, including CED-10/Rac, RhoG/MIG-2, and CDC-42, and downstream mediators, which include the WAVE and WASP regulatory complexes (Walck-Shannon et al., 2016; Walck-Shannon et al., 2015).
We sought to identify additional actin regulators that modulate protrusive activity in dorsal cells. A promising candidate is MIG-10, the single MIG-10/RIAM/Lamellipodin (MRL) family member in C. elegans. As the gene name implies, MIG-10 was originally identified in classic genetic screens as a modulator of cell migration (Manser and Wood, 1990). The canonical mutation, mig-10(ct41), results in an amber stop in Exon 3, resulting in predicted severe truncation and strong loss of function of both MIG-10 isoforms (Manser et al., 1997). In neurons, MIG-10 binds activated CED-10/Rac, which polarizes it to the leading edge of growth cones and is necessary for proper axon guidance (Quinn et al., 2008).
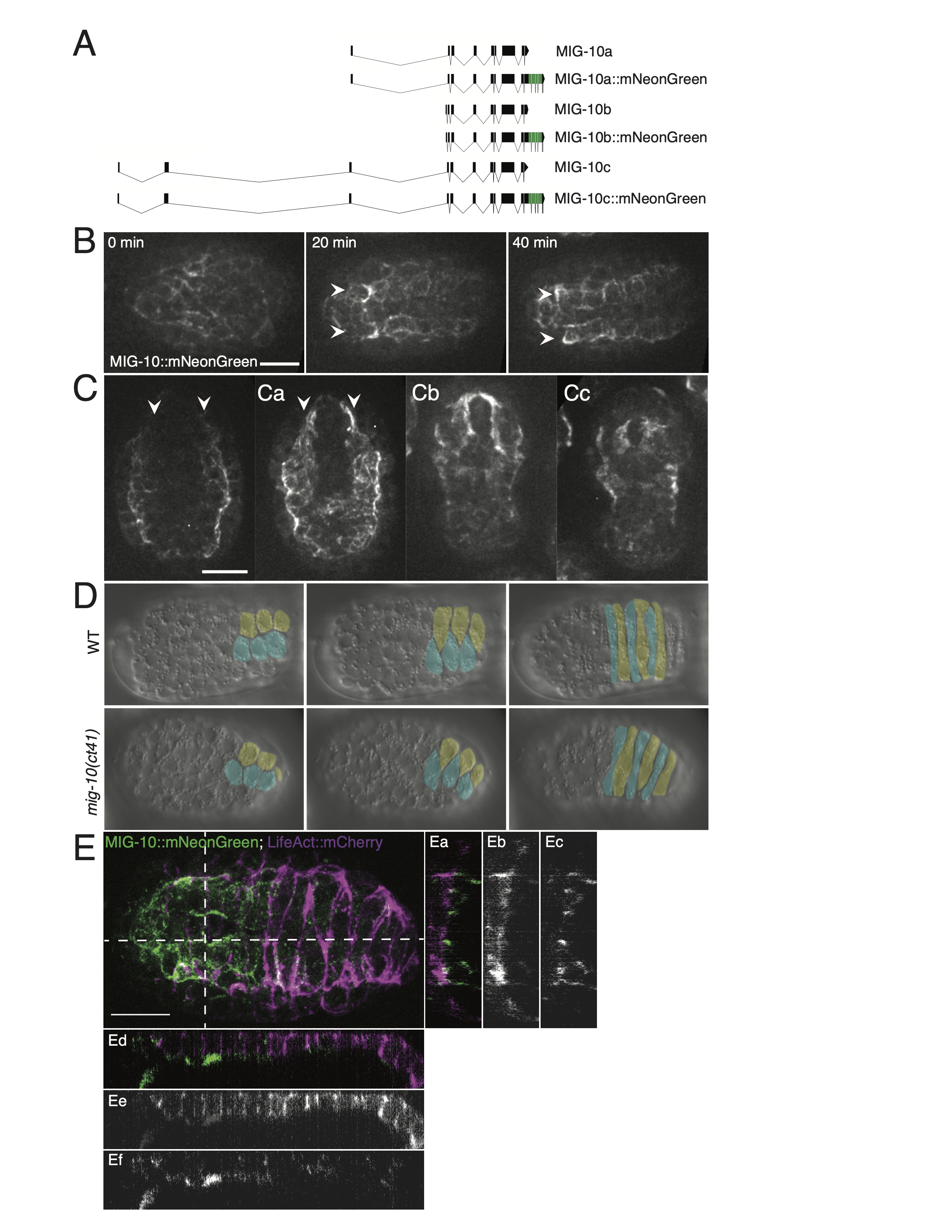
Given established requirements for MIG-10 in cell movement and its connection to Rac signaling, we investigated whether MIG-10 plays a significant role during dorsal intercalation in two ways: (1) by endogenously tagging all mig-10 isoforms with mNeonGreen, and (2) by analyzing mig-10 mutants using 4-dimensional DIC microscopy. We endogenously tagged MIG-10 using a C-terminal insertion in the last exon, which is shared by all mig-10 isoforms (Fig. 1A). The endogenous insertion maintains full function of MIG-10, since we did not observe any Mig phenotypes in embryos, larvae, and adults homozygous for the endogenously tagged version of the gene. The earliest prominent expression of MIG-10 we observed was during early morphogenesis in ventral neuroblasts and muscle precursors. The latter organize into four quadrants, two dorsal and two ventral, as embryogenesis proceeds (Viveiros et al., 2011). The two dorsal muscle quadrants lie immediately ventral to the dorsal epidermis, and show prominent expression of MIG-10::mNG (Fig. 1B, 1C). While it is possible that there is low-level expression of mig-10 in dorsal epidermal cells, if it is expressed there it is present at much lower levels than in dorsal muscle quadrants. We did not see any overlap between epidermally driven LifeAct::mCherry and MIG-10::mNeonGreen (Fig. 1E), further suggesting that detectable dorsal mig-10 expression is restricted to muscle precursors at this stage of development.
The relative lack of expression of MIG-10 in the dorsal epidermis suggests that it is not a major contributor to actin dynamics during dorsal intercalation. We confirmed this using 4d DIC microscopy. mig-10(ct41) homozygotes complete dorsal intercalation in a manner indistinguishable from wild-type embryos (Fig. 1D). In 8 mounts with 21 dorsal presenting embryos, we observed no dorsal intercalation defects in the mig-10(ct41) background, a result indistinguishable from wild-type embryos (no defects in 7 mounts with 26 dorsal presenting wild-type embryos).
Taken together, these results suggest that MIG-10 is not an essential actin regulator in dorsal epidermal cells during dorsal intercalation. It will be interesting to determine if MIG-10 acts redundantly with other actin regulators or if it plays more prominent roles during other cell migration events in the embryo.
Methods
Request a detailed protocolStrains
Hermaphrodite worms were maintained at 20°C on NGM plates with OP50. Strains used: Bristol (N2), SU875 mig-10(jc53[mig-10::mNeonGreen::3xFlag + LoxP]III, SU876 (derived from strain BW315, mig-10(ct41) 3x outcrossed into N2). SU1052 mig-10(jc53[mig-10::mNeonGreen::3xFlag + LoxP]III; curIs11[Plin-26::LifeAct::mCherry::unc-54 3’UTR; unc-119 (+)].
Confocal imaging
MIG-10::mNeonGreen embryos were dissected from adult hermaphrodites and mounted onto 10% agar pads in M9 solution and imaged essentially as described (Zaidel-Bar et al., 2010). Spinning-disc confocal images were acquired with a Z-slice spacing of 0.5 μm using µManager software v1.4.18 (Edelstein et al., 2014) using a Nikon Eclipse E600 microscope controlled via a Prior Z motor (Prior Scientific Instruments, Rockland, Massachusetts) connected to a Yokogawa CSU10 spinning disk scanhead (originally purchased from Perkin-Elmer Life Sciences), a Vortran solid state laser launch controlled using an Arduino control interface (purchased from BioVision Technologies, Exton, Pennsylvania), and a Hamamatsu ORCA-ER charge-coupled device (CCD) camera (Hamamastu Photonics USA, Bridgewater, New Jersey).
4d DIC analysis
Embryos were dissected from adult hermaphrodites and mounted onto 10% agar pads in M9 solution and imaged as described (Walck-Shannon et al., 2015). Image stacks were obtained at 90s intervals. Embryos were scored for dorsal intercalation defects (ipsilateral comigration and failure to intercalate) among the posterior cells of the dorsal array. Color overlays were produced in ImageJ as described (Walck-Shannon et al., 2015).
Genomic Diagrams
Exon-intron diagrams were made using the Exon-Intron Graphic Maker designed and hosted by Nikhil Bhatla. This tool can be accessed at wormweb.org/exonintron
CRISPR
To generate the MIG-10::mNeonGreen knock-in line, we used a plasmid-based self-excising cassette methodology that has been described previously (Dickinson et al., 2015). To generate the appropriate repair template, pJMS5, Gibson cloning was used to replace ccdB sequences in pDD268 with a 5’ homology arm that was approximately 433bp upstream of the Stop codon and a 3’ homology arm beginning with the stop codon and continuing downstream for approximately 538bp amplified from single-worm lysates. The homology arms flanked a self-excising cassette (SEC) which contains a dominant rol allele of sqt-1, a heat shock inducible Cre recombinase and a hygromycin drug resistance marker. pDD268 was generously provided by Dan Dickinson. The guide sequence was cloned into pJW1219 to make pJMS4. pJW1219 was a gift from Jordan Ward (Addgene plasmid # 61250 ; http://n2t.net/addgene:61250 ; RRID:Addgene_61250) (Ward, 2015). pJMS5 was injected at 15 ng/ml into N2 worms with pJMS4 (50ng/ml; pJW1219 carrying the guide sequence for the C-terminal end of mig-10 and Cas9) and co-injection markers pGH8 (Prab-3::mCherry, 10 ng/ml), pCFJ104 (5 ng/ml, Pmyo-3::mCherry), and pCFJ90 (Pmyo-2::mCherry, 2.5 ng/ml). Following selection for rolling worms that no longer carried the extra-chromosomal array, we selected for homozygous rolling worms. These worms were then heat shocked at 32°C for 4 hours to remove the SEC, which left a LoxP site in a synthetic intron between the mNeonGreen and 3xFlag tags.
Sequences for homology arm amplification primers and the sgRNA were as follows:
5’ homology arm Forward primer- 5’-gccaacaatctcatccatctcg-3’
5’ homology arm reverse primer- 5’-acactccatggttgccattttctc-3’
3’ homology arm forward primer-5’-tagacaacatttagaatactggata-3’
3’ homology arm reverse primer-5’-tgtatgcaactgtggaatat-3’
sgRNA 5’-aaatgttgtctaacactcca–3’
References
Funding
This work was funded by NIH grant R01 GM127687 awarded to J.H. J.S. was supported in part by T32 training grant GM007133 through the University of Wisconsin-Madison Program in Genetics.
Reviewed By
Christopher QuinnHistory
Received: December 23, 2021Revision received: January 25, 2022
Accepted: January 27, 2022
Published: February 3, 2022
Copyright
© 2022 by the authors. This is an open-access article distributed under the terms of the Creative Commons Attribution 4.0 International (CC BY 4.0) License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.Citation
Serre, JM; Hardin, J (2022). The Lamellipodin homologue MIG-10 is not essential for dorsal intercalation in the embryonic epidermis of the C. elegans embryo. microPublication Biology. 10.17912/micropub.biology.000522.Download: RIS BibTeX